A burst of ABC genes in the genome of the polyphagous spider mite Tetranychus urticae
- PMID: 23663308
- PMCID: PMC3724490
- DOI: 10.1186/1471-2164-14-317
A burst of ABC genes in the genome of the polyphagous spider mite Tetranychus urticae
Abstract
Background: The ABC (ATP-binding cassette) gene superfamily is widespread across all living species. The majority of ABC genes encode ABC transporters, which are membrane-spanning proteins capable of transferring substrates across biological membranes by hydrolyzing ATP. Although ABC transporters have often been associated with resistance to drugs and toxic compounds, within the Arthropoda ABC gene families have only been characterized in detail in several insects and a crustacean. In this study, we report a genome-wide survey and expression analysis of the ABC gene superfamily in the spider mite, Tetranychus urticae, a chelicerate ~ 450 million years diverged from other Arthropod lineages. T. urticae is a major agricultural pest, and is among of the most polyphagous arthropod herbivores known. The species resists a staggering array of toxic plant secondary metabolites, and has developed resistance to all major classes of pesticides in use for its control.
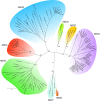
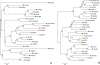
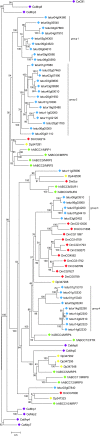
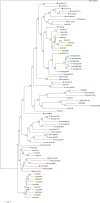
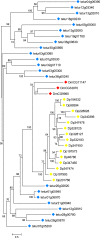
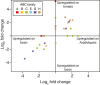
Results: We identified 103 ABC genes in the T. urticae genome, the highest number discovered in a metazoan species to date. Within the T. urticae ABC gene set, all members of the eight currently described subfamilies (A to H) were detected. A phylogenetic analysis revealed that the high number of ABC genes in T. urticae is due primarily to lineage-specific expansions of ABC genes within the ABCC, ABCG and ABCH subfamilies. In particular, the ABCC subfamily harbors the highest number of T. urticae ABC genes (39). In a comparative genomic analysis, we found clear orthologous relationships between a subset of T. urticae ABC proteins and ABC proteins in both vertebrates and invertebrates known to be involved in fundamental cellular processes. These included members of the ABCB-half transporters, and the ABCD, ABCE and ABCF families. Furthermore, one-to-one orthologues could be distinguished between T. urticae proteins and human ABCC10, ABCG5 and ABCG8, the Drosophila melanogaster sulfonylurea receptor and ecdysone-regulated transporter E23. Finally, expression profiling revealed that ABC genes in the ABCC, ABCG ABCH subfamilies were differentially expressed in multi-pesticide resistant mite strains and/or in mites transferred to challenging (toxic) host plants.
Conclusions: In this study we present the first comprehensive analysis of ABC genes in a polyphagous arthropod herbivore. We demonstrate that the broad plant host range and high levels of pesticide resistance in T. urticae are associated with lineage-specific expansions of ABC genes, many of which respond transcriptionally to xenobiotic exposure. This ABC catalogue will serve as a basis for future biochemical and toxicological studies. Obtaining functional evidence that these ABC subfamilies contribute to xenobiotic tolerance should be the priority of future research.
Figures

Similar articles
-
The ABC transporter gene family of Daphnia pulex.BMC Genomics. 2009 Apr 21;10:170. doi: 10.1186/1471-2164-10-170. BMC Genomics. 2009. PMID: 19383151 Free PMC article.
-
Genome-wide identification of ATP-binding cassette (ABC) transporters and conservation of their xenobiotic transporter function in the monogonont rotifer (Brachionus koreanus).Comp Biochem Physiol Part D Genomics Proteomics. 2017 Mar;21:17-26. doi: 10.1016/j.cbd.2016.10.003. Epub 2016 Oct 24. Comp Biochem Physiol Part D Genomics Proteomics. 2017. PMID: 27835832
-
Transcriptome-based identification of ABC transporters in the western tarnished plant bug Lygus hesperus.PLoS One. 2014 Nov 17;9(11):e113046. doi: 10.1371/journal.pone.0113046. eCollection 2014. PLoS One. 2014. PMID: 25401762 Free PMC article.
-
The ABC gene family in arthropods: comparative genomics and role in insecticide transport and resistance.Insect Biochem Mol Biol. 2014 Feb;45:89-110. doi: 10.1016/j.ibmb.2013.11.001. Epub 2013 Nov 28. Insect Biochem Mol Biol. 2014. PMID: 24291285 Review.
-
Can Plant Defence Mechanisms Provide New Approaches for the Sustainable Control of the Two-Spotted Spider Mite Tetranychus urticae?Int J Mol Sci. 2018 Feb 21;19(2):614. doi: 10.3390/ijms19020614. Int J Mol Sci. 2018. PMID: 29466295 Free PMC article. Review.
Cited by
-
Genome streamlining in a minute herbivore that manipulates its host plant.Elife. 2020 Oct 23;9:e56689. doi: 10.7554/eLife.56689. Elife. 2020. PMID: 33095158 Free PMC article.
-
Evolution of chemosensory and detoxification gene families across herbivorous Drosophilidae.bioRxiv [Preprint]. 2023 Mar 16:2023.03.16.532987. doi: 10.1101/2023.03.16.532987. bioRxiv. 2023. Update in: G3 (Bethesda). 2023 Aug 9;13(8):jkad133. doi: 10.1093/g3journal/jkad133. PMID: 36993186 Free PMC article. Updated. Preprint.
-
Evolution of chemosensory and detoxification gene families across herbivorous Drosophilidae.G3 (Bethesda). 2023 Aug 9;13(8):jkad133. doi: 10.1093/g3journal/jkad133. G3 (Bethesda). 2023. PMID: 37317982 Free PMC article.
-
Transcriptome Analysis of the Carmine Spider Mite, Tetranychus cinnabarinus (Boisduval, 1867) (Acari: Tetranychidae), and Its Response to β-Sitosterol.Biomed Res Int. 2015;2015:794718. doi: 10.1155/2015/794718. Epub 2015 May 11. Biomed Res Int. 2015. PMID: 26078964 Free PMC article.
-
The Salivary Protein Repertoire of the Polyphagous Spider Mite Tetranychus urticae: A Quest for Effectors.Mol Cell Proteomics. 2016 Dec;15(12):3594-3613. doi: 10.1074/mcp.M116.058081. Epub 2016 Oct 4. Mol Cell Proteomics. 2016. PMID: 27703040 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

