A detergent-free strategy for the reconstitution of active enzyme complexes from native biological membranes into nanoscale discs
- PMID: 23663692
- PMCID: PMC3702409
- DOI: 10.1186/1472-6750-13-41
A detergent-free strategy for the reconstitution of active enzyme complexes from native biological membranes into nanoscale discs
Abstract
Background: The reconstitution of membrane proteins and complexes into nanoscale lipid bilayer structures has contributed significantly to biochemical and biophysical analyses. Current methods for performing such reconstitutions entail an initial detergent-mediated step to solubilize and isolate membrane proteins. Exposure to detergents, however, can destabilize many membrane proteins and result in a loss of function. Amphipathic copolymers have recently been used to stabilize membrane proteins and complexes following suitable detergent extraction. However, the ability of these copolymers to extract proteins directly from native lipid bilayers for subsequent reconstitution and characterization has not been explored.
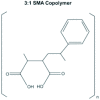
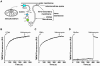
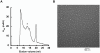
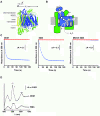
Results: The styrene-maleic acid (SMA) copolymer effectively solubilized membranes of isolated mitochondria and extracted protein complexes. Membrane complexes were reconstituted into polymer-bound nanoscale discs along with endogenous lipids. Using respiratory Complex IV as a model, these particles were shown to maintain the enzymatic activity of multicomponent electron transporting complexes.
Conclusions: We report a novel process for reconstituting fully operational protein complexes directly from cellular membranes into nanoscale lipid bilayers using the SMA copolymer. This facile, single-step strategy obviates the requirement for detergents and yields membrane complexes suitable for structural and functional studies.
Figures

References
-
- Lee AG. How lipids affect the activities of integral membrane proteins. Biochim Biophys Acta. 2004;1666(1–2):62–87. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

