Trip12, a HECT domain E3 ubiquitin ligase, targets Sox6 for proteasomal degradation and affects fiber type-specific gene expression in muscle cells
- PMID: 23663701
- PMCID: PMC3666947
- DOI: 10.1186/2044-5040-3-11
Trip12, a HECT domain E3 ubiquitin ligase, targets Sox6 for proteasomal degradation and affects fiber type-specific gene expression in muscle cells
Abstract
Background: A sophisticated level of coordinated gene expression is necessary for skeletal muscle fibers to obtain their unique functional identities. We have previously shown that the transcription factor Sox6 plays an essential role in coordinating muscle fiber type differentiation by acting as a transcriptional suppressor of slow fiber-specific genes. Currently, mechanisms regulating the activity of Sox6 in skeletal muscle and how these mechanisms affect the fiber phenotype remain unknown.
Methods: Yeast two-hybrid screening was used to identify binding partners of Sox6 in muscle. Small interfering RNA (siRNA)-mediated knockdown of one of the Sox6 binding proteins, Trip12, was used to determine its effect on Sox6 activity in C2C12 myotubes using quantitative analysis of fiber type-specific gene expression.
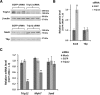
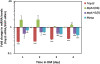
Results: We found that the E3 ligase Trip12, a HECT domain E3 ubiquitin ligase, recognizes and polyubiquitinates Sox6. Inhibiting Trip12 or the 26S proteasome activity resulted in an increase in Sox6 protein levels in C2C12 myotubes. This control of Sox6 activity in muscle cells via Trip12 ubiquitination has significant phenotypic outcomes. Knockdown of Trip12 in C2C12 myotubes led to upregulation of Sox6 protein levels and concurrently to a decrease in slow fiber-specific Myh7 expression coupled with an increased expression in fast fiber-specific Myh4. Therefore, regulation of Sox6 cellular levels by the ubiquitin-proteasome system can induce identity-changing alterations in the expression of fiber type-specific genes in muscle cells.
Conclusions: Based on our data, we propose that in skeletal muscle, E3 ligases have a significant role in regulating fiber type-specific gene expression, expanding their importance in muscle beyond their well-established role in atrophy.
Figures
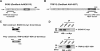

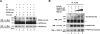

References
-
- Seene T, Kaasik P, Umnova M. Structural rearrangements in contractile apparatus and resulting skeletal muscle remodelling: effect of exercise training. J Sports Med Phys Fitness. 2009;49:410–423. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

