Reprogramming the maternal zebrafish genome after fertilization to match the paternal methylation pattern
- PMID: 23663776
- PMCID: PMC4030421
- DOI: 10.1016/j.cell.2013.04.030
Reprogramming the maternal zebrafish genome after fertilization to match the paternal methylation pattern
Abstract
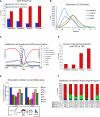
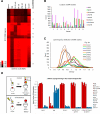
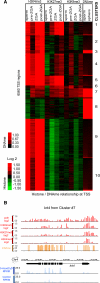
Early vertebrate embryos must achieve totipotency and prepare for zygotic genome activation (ZGA). To understand this process, we determined the DNA methylation (DNAme) profiles of zebrafish gametes, embryos at different stages, and somatic muscle and compared them to gene activity and histone modifications. Sperm chromatin patterns are virtually identical to those at ZGA. Unexpectedly, the DNA of many oocyte genes important for germline functions (i.e., piwil1) or early development (i.e., hox genes) is methylated, but the loci are demethylated during zygotic cleavage stages to precisely the state observed in sperm, even in parthenogenetic embryos lacking a replicating paternal genome. Furthermore, this cohort constitutes the genes and loci that acquire DNAme during development (i.e., ZGA to muscle). Finally, DNA methyltransferase inhibition experiments suggest that DNAme silences particular gene and chromatin cohorts at ZGA, preventing their precocious expression. Thus, zebrafish achieve a totipotent chromatin state at ZGA through paternal genome competency and maternal genome DNAme reprogramming.
Copyright © 2013 Elsevier Inc. All rights reserved.
Figures



Comment in
-
Beyond DNA: programming and inheritance of parental methylomes.Cell. 2013 May 9;153(4):737-9. doi: 10.1016/j.cell.2013.04.044. Cell. 2013. PMID: 23663772 Free PMC article.
-
Development: Zebrafish early methylomes.Nat Rev Genet. 2013 Jul;14(7):443. doi: 10.1038/nrg3515. Epub 2013 May 29. Nat Rev Genet. 2013. PMID: 23715432 No abstract available.
References
-
- Almeida RD, Loose M, Sottile V, Matsa E, Denning C, Young L, Johnson AD, Gering M, Ruzov A. 5-hydroxymethyl-cytosine enrichment of non-committed cells is not a universal feature of vertebrate development. Epigenetics. 2012;7:383–389. - PubMed
-
- Bogdanovic O, Fernandez-Miñán A, Tena JJ, de la Calle-Mustienes E, Hidalgo C, van Kruysbergen I, van Heeringen SJ, Veenstra GJ, Gómez-Skarmeta JL. Dynamics of enhancer chromatin signatures mark the transition from pluripotency to cell specification during embryogenesis. Genome Res. 2012;22:2043–2053. - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

