The structural basis for the allosteric regulation of ribonucleotide reductase
- PMID: 23663976
- PMCID: PMC4059395
- DOI: 10.1016/B978-0-12-386931-9.00014-3
The structural basis for the allosteric regulation of ribonucleotide reductase
Abstract
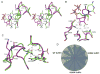
Ribonucleotide reductases (RRs) catalyze a crucial step of de novo DNA synthesis by converting ribonucleoside diphosphates to deoxyribonucleoside diphosphates. Tight control of the dNTP pool is essential for cellular homeostasis. The activity of the enzyme is tightly regulated at the S-phase by allosteric regulation. Recent structural studies by our group and others provided the molecular basis for understanding how RR recognizes substrates, how it interacts with chemotherapeutic agents, and how it is regulated by its allosteric regulators ATP and dATP. This review discusses the molecular basis of allosteric regulation and substrate recognition of RR, and particularly the discovery that subunit oligomerization is an important prerequisite step in enzyme inhibition.
Copyright © 2013 Elsevier Inc. All rights reserved.
Figures
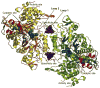



References
-
- Monod J, Changeux JP, Jacob F. Allosteric proteins and cellular control systems. J Mol Biol. 1963;6:306–29. - PubMed
-
- Changeux JP, Edelstein SJ. Allosteric mechanisms of signal transduction. Science. 2005;308(5727):1424–8. - PubMed
-
- Monod J, Wyman J, Changeux JP. On the nature of allosteric transitions: a plausible model. J Mol Biol. 1965;12:88–118. - PubMed
-
- Koshland DE, Jr, Nemethy G, Filmer D. Comparison of experimental binding data and theoretical models in proteins containing subunits. Biochemistry. 1966;5(1):365–85. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

