Viral infection controlled by a calcium-dependent lipid-binding module in ALIX
- PMID: 23664863
- PMCID: PMC4129370
- DOI: 10.1016/j.devcel.2013.04.003
Viral infection controlled by a calcium-dependent lipid-binding module in ALIX
Abstract
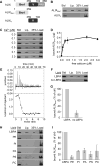
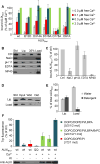
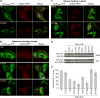
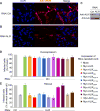
ALIX plays a role in nucleocapsid release during viral infection, as does lysobisphosphatidic acid (LBPA). However, the mechanism remains unclear. Here we report that LBPA is recognized within an exposed site in ALIX Bro1 domain predicted by MODA, an algorithm for discovering membrane-docking areas in proteins. LBPA interactions revealed a strict requirement for a structural calcium tightly bound near the lipid interaction site. Unlike other calcium- and phospholipid-binding proteins, the all-helical triangle-shaped fold of the Bro1 domain confers selectivity for LBPA via a pair of hydrophobic residues in a flexible loop, which undergoes a conformational change upon membrane association. Both LBPA and calcium binding are necessary for endosome association and virus infection, as are ALIX ESCRT binding and dimerization capacity. We conclude that LBPA recruits ALIX onto late endosomes via the calcium-bound Bro1 domain, triggering a conformational change in ALIX to mediate the delivery of viral nucleocapsids to the cytosol during infection.
Copyright © 2013 Elsevier Inc. All rights reserved.
Figures

References
-
- Abagyan R, Totrov M. Biased probability Monte Carlo conformational searches and electrostatic calculations for peptides and proteins. Journal of molecular biology. 1994;235:983–1002. - PubMed
-
- Bordier C. Phase separation of integral membrane proteins in Triton X-114 solution. J Biol Chem. 1981;256:1604–1607. - PubMed
-
- Cabezas A, Bache KG, Brech A, Stenmark H. Alix regulates cortical actin and the spatial distribution of endosomes. J Cell Sci. 2005;118:2625–2635. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

