A transcriptome-wide RNAi screen in the Drosophila ovary reveals factors of the germline piRNA pathway
- PMID: 23665227
- PMCID: PMC3724427
- DOI: 10.1016/j.molcel.2013.04.007
A transcriptome-wide RNAi screen in the Drosophila ovary reveals factors of the germline piRNA pathway
Abstract
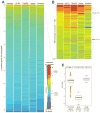
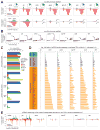
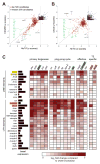
The Drosophila piRNA pathway provides an RNA-based immune system that defends the germline genome against selfish genetic elements. Two interrelated branches of the piRNA system exist: somatic cells that support oogenesis only employ Piwi, whereas germ cells utilize a more elaborate pathway centered on the three gonad-specific Argonaute proteins (Piwi, Aubergine, and Argonaute 3). While several key factors of each branch have been identified, our current knowledge is insufficient to explain the complex workings of the piRNA machinery. Here, we report a reverse genetic screen spanning the ovarian transcriptome in an attempt to uncover the full repertoire of genes required for piRNA-mediated transposon silencing in the female germline. Our screen reveals key factors of piRNA-mediated transposon silencing, including the piRNA biogenesis factors CG2183 (GASZ) and Deadlock. Our data uncover a previously unanticipated set of factors preferentially required for repression of different transposon types.
Copyright © 2013 Elsevier Inc. All rights reserved.
Figures
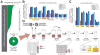


References
-
- Aravin A, Gaidatzis D, Pfeffer S, Lagos-Quintana M, Landgraf P, Iovino N, Morris P, Brownstein MJ, Kuramochi-Miyagawa S, Nakano T, et al. A novel class of small RNAs bind to MILI protein in mouse testes. Nature. 2006;442:203–207. - PubMed
-
- Brennecke J, Aravin AA, Stark A, Dus M, Kellis M, Sachidanandam R, Hannon GJ. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell. 2007;128:1089–1103. - PubMed
-
- Chang YF, Imam JS, Wilkinson MF. The nonsense-mediated decay RNA surveillance pathway. Annu Rev Biochem. 2007;76:51–74. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

