GAGE-B: an evaluation of genome assemblers for bacterial organisms
- PMID: 23665771
- PMCID: PMC3702249
- DOI: 10.1093/bioinformatics/btt273
GAGE-B: an evaluation of genome assemblers for bacterial organisms
Abstract
Motivation: A large and rapidly growing number of bacterial organisms have been sequenced by the newest sequencing technologies. Cheaper and faster sequencing technologies make it easy to generate very high coverage of bacterial genomes, but these advances mean that DNA preparation costs can exceed the cost of sequencing for small genomes. The need to contain costs often results in the creation of only a single sequencing library, which in turn introduces new challenges for genome assembly methods.
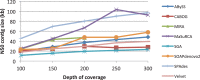
Results: We evaluated the ability of multiple genome assembly programs to assemble bacterial genomes from a single, deep-coverage library. For our comparison, we chose bacterial species spanning a wide range of GC content and measured the contiguity and accuracy of the resulting assemblies. We compared the assemblies produced by this very high-coverage, one-library strategy to the best assemblies created by two-library sequencing, and we found that remarkably good bacterial assemblies are possible with just one library. We also measured the effect of read length and depth of coverage on assembly quality and determined the values that provide the best results with current algorithms.
Contact: salzberg@jhu.edu
Supplementary information: Supplementary data are available at Bioinformatics online.
Figures
References
-
- Aronesty E. Ea-utils: command-line tools for processing biological sequencing data. 2011. http://code.google.com/p/ea-utils (3 June 2013, date last accessed)
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous