Nuclear magnetic resonance spectroscopy of the circadian clock of cyanobacteria
- PMID: 23667047
- PMCID: PMC6296334
- DOI: 10.1093/icb/ict054
Nuclear magnetic resonance spectroscopy of the circadian clock of cyanobacteria
Abstract
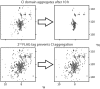
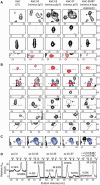
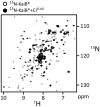
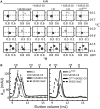
The most well-understood circadian clock at the level of molecular mechanisms is that of cyanobacteria. This overview is on how solution-state nuclear magnetic resonance (NMR) spectroscopy has contributed to this understanding. By exciting atomic spin-½ nuclei in a strong magnetic field, NMR obtains information on their chemical environments, inter-nuclear distances, orientations, and motions. NMR protein samples are typically aqueous, often at near-physiological pH, ionic strength, and temperature. The level of information obtainable by NMR depends on the quality of the NMR sample, by which we mean the solubility and stability of proteins. Here, we use examples from our laboratory to illustrate the advantages and limitations of the technique.
Figures


Similar articles
-
Orderly wheels of the cyanobacterial clock.Proc Natl Acad Sci U S A. 2012 Oct 16;109(42):16760-1. doi: 10.1073/pnas.1214901109. Epub 2012 Oct 8. Proc Natl Acad Sci U S A. 2012. PMID: 23045681 Free PMC article. No abstract available.
-
Low temperature nullifies the circadian clock in cyanobacteria through Hopf bifurcation.Proc Natl Acad Sci U S A. 2017 May 30;114(22):5641-5646. doi: 10.1073/pnas.1620378114. Epub 2017 May 17. Proc Natl Acad Sci U S A. 2017. PMID: 28515313 Free PMC article.
-
Structure, function, and mechanism of the core circadian clock in cyanobacteria.J Biol Chem. 2018 Apr 6;293(14):5026-5034. doi: 10.1074/jbc.TM117.001433. Epub 2018 Feb 13. J Biol Chem. 2018. PMID: 29440392 Free PMC article. Review.
-
Timing the day: what makes bacterial clocks tick?Nat Rev Microbiol. 2017 Apr;15(4):232-242. doi: 10.1038/nrmicro.2016.196. Epub 2017 Feb 20. Nat Rev Microbiol. 2017. PMID: 28216658 Free PMC article. Review.
-
Diversity of KaiC-based timing systems in marine Cyanobacteria.Mar Genomics. 2014 Apr;14:3-16. doi: 10.1016/j.margen.2013.12.006. Epub 2014 Jan 3. Mar Genomics. 2014. PMID: 24388874 Review.
References
-
- Akiyama S, Nohara A, Ito K, Maeda Y. Assembly and disassembly dynamics of the cyanobacterial periodosome. Mol Cell. 2008;29:703–16. - PubMed
-
- Amero C, Asuncion Dura M, Noirclerc-Savoye M, Perollier A, Gallet B, Plevin M, Vernet T, Franzetti B, Boisbouvier J. A systematic mutagenesis-driven strategy for site-resolved NMR studies of supramolecular assemblies. J Biomol NMR. 2011;50:229–36. - PubMed
-
- Ayala I, Sounier R, Use N, Gans P, Boisbouvier J. An efficient protocol for the complete incorporation of methyl-protonated alanine in perdeuterated protein. J Biomol NMR. 2009;43:111–9. - PubMed
-
- Bass J. Circadian topology of metabolism. Nature. 2012;491:348–56. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

