Viral antigen induces differentiation of Foxp3+ natural regulatory T cells in influenza virus-infected mice
- PMID: 23667113
- PMCID: PMC3703618
- DOI: 10.4049/jimmunol.1203302
Viral antigen induces differentiation of Foxp3+ natural regulatory T cells in influenza virus-infected mice
Abstract
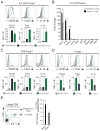
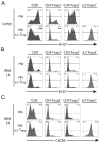
We examined the formation, participation, and functional specialization of virus-reactive Foxp3(+) regulatory T cells (Tregs) in a mouse model of influenza virus infection. "Natural" Tregs generated intrathymically, based on interactions with a self-peptide, proliferated in response to a homologous viral Ag in the lungs and, to a lesser extent, in the lung-draining mediastinal lymph nodes (medLNs) of virus-infected mice. In contrast, conventional CD4(+) T cells with identical TCR specificity underwent little or no conversion to become "adaptive" Tregs. The virus-reactive Tregs in the medLNs and the lungs of infected mice upregulated a variety of molecules associated with Treg activation, as well as acquired expression of molecules (T-bet, Blimp-1, and IL-10) that confer functional specialization to Tregs. Notably, however, the phenotypes of the T-bet(+) Tregs obtained from these sites were distinct, because Tregs isolated from the lungs expressed significantly higher levels of T-bet, Blimp-1, and IL-10 than did Tregs from the medLNs. Adoptive transfer of Ag-reactive Tregs led to decreased proliferation of antiviral CD4(+) and CD8(+) effector T cells in the lungs of infected hosts, whereas depletion of Tregs had a reciprocal effect. These studies demonstrate that thymically generated Tregs can become activated by a pathogen-derived peptide and acquire discrete T-bet(+) Treg phenotypes while participating in and modulating an antiviral immune response.
Conflict of interest statement
The authors declare no financial conflicts of interest.
Figures
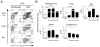



References
-
- Shevach EM. Biological functions of regulatory T cells. Adv Immunol. 2011;112:137–176. - PubMed
-
- Sakaguchi S, Sakaguchi N, Asano M, Itoh M, Toda M. Immunologic self-tolerance maintained by activated T cells expressing IL-2 receptor alpha-chains (CD25). Breakdown of a single mechanism of self-tolerance causes various autoimmune diseases. J Immunol. 1995;155:1151–1164. - PubMed
-
- Kim JM, Rasmussen JP, Rudensky AY. Regulatory T cells prevent catastrophic autoimmunity throughout the lifespan of mice. Nat Immunol. 2007;8:191–197. - PubMed
-
- Belkaid Y, Tarbell K. Regulatory T cells in the control of host-microorganism interactions. Annu Rev Immunol. 2009;27:551–589. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

