Advanced proteomic analyses yield a deep catalog of ubiquitylation targets in Arabidopsis
- PMID: 23667124
- PMCID: PMC3694690
- DOI: 10.1105/tpc.112.108613
Advanced proteomic analyses yield a deep catalog of ubiquitylation targets in Arabidopsis
Abstract
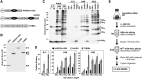
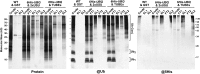
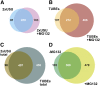
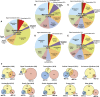
The posttranslational addition of ubiquitin (Ub) profoundly controls the half-life, interactions, and/or trafficking of numerous intracellular proteins. Using stringent two-step affinity methods to purify Ub-protein conjugates followed by high-sensitivity mass spectrometry, we identified almost 950 ubiquitylation substrates in whole Arabidopsis thaliana seedlings. The list includes key factors regulating a wide range of biological processes, including metabolism, cellular transport, signal transduction, transcription, RNA biology, translation, and proteolysis. The ubiquitylation state of more than half of the targets increased after treating seedlings with the proteasome inhibitor MG132 (carbobenzoxy-Leu-Leu-Leu-al), strongly suggesting that Ub addition commits many to degradation by the 26S proteasome. Ub-attachment sites were resolved for a number of targets, including six of the seven Lys residues on Ub itself with a Lys-48>Lys-63>Lys-11>>>Lys-33/Lys-29/Lys-6 preference. However, little sequence consensus was detected among conjugation sites, indicating that the local environment has little influence on global ubiquitylation. Intriguingly, the level of Lys-11-linked Ub polymers increased substantially upon MG132 treatment, revealing that they might be important signals for proteasomal breakdown. Taken together, this proteomic analysis illustrates the breadth of plant processes affected by ubiquitylation and provides a deep data set of individual targets from which to explore the roles of Ub in various physiological and developmental pathways.
Figures

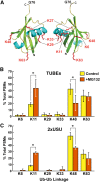
References
-
- Baunsgaard L., Fuglsang A.T., Jahn T., Korthout H.A., de Boer A.H., Palmgren M.G. (1998). The 14-3-3 proteins associate with the plant plasma membrane H(+)-ATPase to generate a fusicoccin binding complex and a fusicoccin responsive system. Plant J. 13: 661–671 - PubMed
-
- Book A.J., Smalle J., Lee K.H., Yang P., Walker J.M., Casper S., Holmes J.H., Russo L.A., Buzzinotti Z.W., Jenik P.D., Vierstra R.D. (2009). The RPN5 subunit of the 26S proteasome is essential for gametogenesis, sporophyte development, and complex assembly in Arabidopsis. Plant Cell 21: 460–478 - PMC - PubMed
-
- Chevalier D., Walker J.C. (2005). Functional genomics of protein kinases in plants. Brief. Funct. Genomics Proteomics 3: 362–371 - PubMed
-
- Colaert N., Helsens K., Martens L., Vandekerckhove J., Gevaert K. (2009). Improved visualization of protein consensus sequences by iceLogo. Nat. Methods 6: 786–787 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

