LipidHome: a database of theoretical lipids optimized for high throughput mass spectrometry lipidomics
- PMID: 23667450
- PMCID: PMC3646891
- DOI: 10.1371/journal.pone.0061951
LipidHome: a database of theoretical lipids optimized for high throughput mass spectrometry lipidomics
Abstract
Protein sequence databases are the pillar upon which modern proteomics is supported, representing a stable reference space of predicted and validated proteins. One example of such resources is UniProt, enriched with both expertly curated and automatic annotations. Taken largely for granted, similar mature resources such as UniProt are not available yet in some other "omics" fields, lipidomics being one of them. While having a seasoned community of wet lab scientists, lipidomics lies significantly behind proteomics in the adoption of data standards and other core bioinformatics concepts. This work aims to reduce the gap by developing an equivalent resource to UniProt called 'LipidHome', providing theoretically generated lipid molecules and useful metadata. Using the 'FASTLipid' Java library, a database was populated with theoretical lipids, generated from a set of community agreed upon chemical bounds. In parallel, a web application was developed to present the information and provide computational access via a web service. Designed specifically to accommodate high throughput mass spectrometry based approaches, lipids are organised into a hierarchy that reflects the variety in the structural resolution of lipid identifications. Additionally, cross-references to other lipid related resources and papers that cite specific lipids were used to annotate lipid records. The web application encompasses a browser for viewing lipid records and a 'tools' section where an MS1 search engine is currently implemented. LipidHome can be accessed at http://www.ebi.ac.uk/apweiler-srv/lipidhome.
Conflict of interest statement
Figures

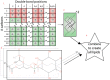
References
-
- Orešič M, Hänninen V, Vidal-Puig A (2013) Lipidomics: A New Window to Biomedical Frontiers. Science and Nature 2(1): 26–31. - PubMed
-
- Hu C, Kong H, Qu F, Li Y, Yu Z, et al. (2011) Application of plasma lipidomics in studying the response of patients with essential hypertension to antihypertensive drug therapy. Molecular bioSystems 7: 3271–3279. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

