Diversity of T cell epitopes in Plasmodium falciparum circumsporozoite protein likely due to protein-protein interactions
- PMID: 23667476
- PMCID: PMC3646838
- DOI: 10.1371/journal.pone.0062427
Diversity of T cell epitopes in Plasmodium falciparum circumsporozoite protein likely due to protein-protein interactions
Abstract
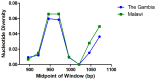
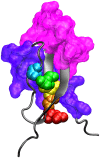
Circumsporozoite protein (CS) is a leading vaccine antigen for falciparum malaria, but is highly polymorphic in natural parasite populations. The factors driving this diversity are unclear, but non-random assortment of the T cell epitopes TH2 and TH3 has been observed in a Kenyan parasite population. The recent publication of the crystal structure of the variable C terminal region of the protein allows the assessment of the impact of diversity on protein structure and T cell epitope assortment. Using data from the Gambia (55 isolates) and Malawi (235 isolates), we evaluated the patterns of diversity within and between epitopes in these two distantly-separated populations. Only non-synonymous mutations were observed with the vast majority in both populations at similar frequencies suggesting strong selection on this region. A non-random pattern of T cell epitope assortment was seen in Malawi and in the Gambia, but structural analysis indicates no intramolecular spatial interactions. Using the information from these parasite populations, structural analysis reveals that polymorphic amino acids within TH2 and TH3 colocalize to one side of the protein, surround, but do not involve, the hydrophobic pocket in CS, and predominately involve charge switches. In addition, free energy analysis suggests residues forming and behind the novel pocket within CS are tightly constrained and well conserved in all alleles. In addition, free energy analysis shows polymorphic residues tend to be populated by energetically unfavorable amino acids. In combination, these findings suggest the diversity of T cell epitopes in CS may be primarily an evolutionary response to intermolecular interactions at the surface of the protein potentially counteracting antibody-mediated immune recognition or evolving host receptor diversity.
Conflict of interest statement
Figures






References
-
- Waitumbi JN, Anyona SB, Hunja CW, Kifude CM, Polhemus ME, et al. (2009) Impact of RTS,S/AS02(A) and RTS,S/AS01(B) on genotypes of P. falciparum in adults participating in a malaria vaccine clinical trial. PLoS ONE 4: e7849 doi:10.1371/journal.pone.0007849. - DOI - PMC - PubMed
-
- Agnandji ST, Lell B, Soulanoudjingar SS, Fernandes JF, Abossolo BP, et al. (2011) First results of phase 3 trial of RTS,S/AS01 malaria vaccine in African children. N Engl J Med 365: 1863–1875 doi:10.1056/NEJMoa1102287. - DOI - PubMed
-
- Agnandji ST, Lell B, Fernandes JF, Abossolo BP, Methogo BGNO, et al. (2012) A phase 3 trial of RTS,S/AS01 malaria vaccine in African infants. N Engl J Med 367: 2284–2295 doi:10.1056/NEJMoa1208394. - DOI - PMC - PubMed
-
- Takala SL, Coulibaly D, Thera MA, Batchelor AH, Cummings MP, et al. (2009) Extreme polymorphism in a vaccine antigen and risk of clinical malaria: implications for vaccine development. Sci Transl Med 1: 2ra5 doi:10.1126/scitranslmed.3000257. - DOI - PMC - PubMed
-
- Bailey JA, Mvalo T, Aragam N, Weiser M, Congdon S, et al. (2012) Use of massively parallel pyrosequencing to evaluate the diversity of and selection on Plasmodium falciparum csp T-cell epitopes in Lilongwe, Malawi. J Infect Dis 206: 580–587 doi:10.1093/infdis/jis329. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

