De novo assembly, functional annotation and comparative analysis of Withania somnifera leaf and root transcriptomes to identify putative genes involved in the withanolides biosynthesis
- PMID: 23667511
- PMCID: PMC3648579
- DOI: 10.1371/journal.pone.0062714
De novo assembly, functional annotation and comparative analysis of Withania somnifera leaf and root transcriptomes to identify putative genes involved in the withanolides biosynthesis
Abstract
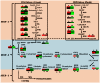
Withania somnifera is one of the most valuable medicinal plants used in Ayurvedic and other indigenous medicine systems due to bioactive molecules known as withanolides. As genomic information regarding this plant is very limited, little information is available about biosynthesis of withanolides. To facilitate the basic understanding about the withanolide biosynthesis pathways, we performed transcriptome sequencing for Withania leaf (101L) and root (101R) which specifically synthesize withaferin A and withanolide A, respectively. Pyrosequencing yielded 8,34,068 and 7,21,755 reads which got assembled into 89,548 and 1,14,814 unique sequences from 101L and 101R, respectively. A total of 47,885 (101L) and 54,123 (101R) could be annotated using TAIR10, NR, tomato and potato databases. Gene Ontology and KEGG analyses provided a detailed view of all the enzymes involved in withanolide backbone synthesis. Our analysis identified members of cytochrome P450, glycosyltransferase and methyltransferase gene families with unique presence or differential expression in leaf and root and might be involved in synthesis of tissue-specific withanolides. We also detected simple sequence repeats (SSRs) in transcriptome data for use in future genetic studies. Comprehensive sequence resource developed for Withania, in this study, will help to elucidate biosynthetic pathway for tissue-specific synthesis of secondary plant products in non-model plant organisms as well as will be helpful in developing strategies for enhanced biosynthesis of withanolides through biotechnological approaches.
Conflict of interest statement
Figures




References
-
- Kaileh M, Berghe WV, Boone E, Essawi T, Haegeman G (2007) Screening of indigenous Palestinian medicinal plants for potential anti-inflammatory and cytotoxic activity. J Ethn Pharmacol 113: 510–516. - PubMed
-
- Standeven R (1998) Withania somnifera. . Eur J Herbal Med 4: 17–22.
-
- Kushwaha S, Soni VK, Singh PK, Bano N, Kumar A, et al. (2012) Withania somnifera chemotypes NMITLI 101R, NMITLI 118R, NMITLI 128R and withaferin A protect Mastomys coucha from Brugia malayi infection. Parasite Immunol 34: 199–209. - PubMed
-
- VenMurthy MR, Ranjekar PK, Ramassamy C, Deshpande M (2010) Scientific basis for the use of Indian Ayurvedic medicinal plants in the treatment of neurodegenerative disorders: Ashwagandha. Cent Nerv Syst Agents Med Chem 10: 238–246. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

