Prdm14 promotes germline fate and naive pluripotency by repressing FGF signalling and DNA methylation
- PMID: 23670199
- PMCID: PMC3701237
- DOI: 10.1038/embor.2013.67
Prdm14 promotes germline fate and naive pluripotency by repressing FGF signalling and DNA methylation
Abstract
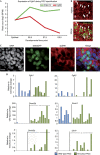
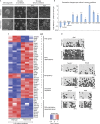
Primordial germ cells (PGCs) and somatic cells originate from postimplantation epiblast cells in mice. As pluripotency is lost upon differentiation of somatic lineages, a naive epigenome and the pluripotency network are re-established during PGC development. Here we demonstrate that Prdm14 contributes not only to PGC specification, but also to naive pluripotency in embryonic stem (ES) cells by repressing the DNA methylation machinery and fibroblast growth factor (FGF) signalling. This indicates a critical role for Prdm14 in programming PGCs and promoting pluripotency in ES cells.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures


References
-
- Ohinata Y et al. (2005) Blimp1 is a critical determinant of the germ cell lineage in mice. Nature 436: 207–213 - PubMed
-
- Weber S et al. (2009) Critical function of AP-2gamma/TCFAP2C in mouse embryonic germ cell maintenance. Biol Reprod 82: 214–223 - PubMed
-
- Yamaji M, Seki Y, Kurimoto K, Yabuta Y, Yuasa M, Shigeta M, Yamanaka K, Ohinata Y, Saitou M (2008) Critical function of Prdm14 for the establishment of the germ cell lineage in mice. Nat Genet 40: 1016–1022 - PubMed
-
- Seki Y, Yamaji M, Yabuta Y, Sano M, Shigeta M, Matsui Y, Saga Y, Tachibana M, Shinkai Y, Saitou M (2007) Cellular dynamics associated with the genome-wide epigenetic reprogramming in migrating primordial germ cells in mice. Development 134: 2627–2638 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

