MoMA-LigPath: a web server to simulate protein-ligand unbinding
- PMID: 23671332
- PMCID: PMC3692135
- DOI: 10.1093/nar/gkt380
MoMA-LigPath: a web server to simulate protein-ligand unbinding
Abstract
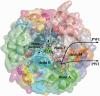
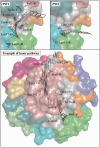
Protein-ligand interactions taking place far away from the active site, during ligand binding or release, may determine molecular specificity and activity. However, obtaining information about these interactions with experimental or computational methods remains difficult. The computational tool presented in this article, MoMA-LigPath, is based on a mechanistic representation of the molecular system, considering partial flexibility, and on the application of a robotics-inspired algorithm to explore the conformational space. Such a purely geometric approach, together with the efficiency of the exploration algorithm, enables the simulation of ligand unbinding within short computing time. Ligand unbinding pathways generated by MoMA-LigPath are a first approximation that can provide useful information about protein-ligand interactions. When needed, this approximation can be subsequently refined and analyzed using state-of-the-art energy models and molecular modeling methods. MoMA-LigPath is available at http://moma.laas.fr. The web server is free and open to all users, with no login requirement.
Figures
References
-
- Chaloupková R, Sýkorová J, Prokop Z, Jesenská A, Monincová M, Pavlová M, Tsuda M, Nagata Y, Damborský J. Modification of activity and specificity of haloalkane dehalogenase from Sphingomonas paucimobilis UT26 by engineering of its entrance tunnel. J. Biol. Chem. 2003;278:52622–52628. - PubMed
-
- Lafaquière V, Barbe S, Puech-Guenot S, Guieysse D, Cortés J, Monsan P, Siméon T, André I, Remaud-Siméon M. Control of lipase enantioselectivity by engineering the substrate binding site and access channel. ChemBioChem. 2009;10:2760–2771. - PubMed
-
- Isralewitz B, Gao M, Schulten K. Steered molecular dynamics and mechanical functions of proteins. Curr. Opin. Struct. Biol. 2001;11:224–230. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

