Differences in gastric carcinoma microenvironment stratify according to EBV infection intensity: implications for possible immune adjuvant therapy
- PMID: 23671415
- PMCID: PMC3649992
- DOI: 10.1371/journal.ppat.1003341
Differences in gastric carcinoma microenvironment stratify according to EBV infection intensity: implications for possible immune adjuvant therapy
Abstract
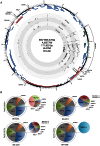
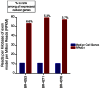
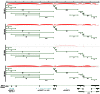
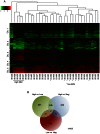
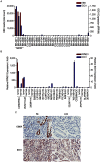
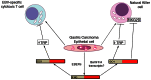
Epstein-Barr virus (EBV) is associated with roughly 10% of gastric carcinomas worldwide (EBVaGC). Although previous investigations provide a strong link between EBV and gastric carcinomas, these studies were performed using selected EBV gene probes. Using a cohort of gastric carcinoma RNA-seq data sets from The Cancer Genome Atlas (TCGA), we performed a quantitative and global assessment of EBV gene expression in gastric carcinomas and assessed EBV associated cellular pathway alterations. EBV transcripts were detected in 17% of samples but these samples varied significantly in EBV coverage depth. In four samples with the highest EBV coverage (hiEBVaGC - high EBV associated gastric carcinoma), transcripts from the BamHI A region comprised the majority of EBV reads. Expression of LMP2, and to a lesser extent, LMP1 were also observed as was evidence of abortive lytic replication. Analysis of cellular gene expression indicated significant immune cell infiltration and a predominant IFNG response in samples expressing high levels of EBV transcripts relative to samples expressing low or no EBV transcripts. Despite the apparent immune cell infiltration, high levels of the cytotoxic T-cell (CTL) and natural killer (NK) cell inhibitor, IDO1, was observed in the hiEBVaGCs samples suggesting an active tolerance inducing pathway in this subgroup. These results were confirmed in a separate cohort of 21 Vietnamese gastric carcinoma samples using qRT-PCR and on tissue samples using in situ hybridization and immunohistochemistry. Lastly, a panel of tumor suppressors and candidate oncogenes were expressed at lower levels in hiEBVaGC versus EBV-low and EBV-negative gastric cancers suggesting the direct regulation of tumor pathways by EBV.
Conflict of interest statement
The authors, Teresa A. Lehman and Michael B. Seddon are currently under the employment of BioServe Biotechnologies, Ltd. BioServe Biotechnologies, Ltd provided funds, resources and equipment to conduct some of the studies and provided salary and benefits for Teresa A. Lehman and Michael B. Seddon. This does not alter our adherence to all PLoS Pathogens policies on sharing data and materials.
Figures




References
-
- Burke A, Yen T, Shekitka K, Sobin L (1990) Lymphoepithelial carcinoma of the stomach with Epstein-Barr virus demonstrated by polymerase chain reaction. Mod Pathol 3: 377–380. - PubMed
-
- Morewaya J, Koriyama C, Akiba S, Shan D, Itoh T, et al. (2004) Epstein-Barr virus-associated gastric carcinoma in Papua New Guinea. Oncol Rep 12: 1093–1098. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

