Experimental interrogation of the path dependence and stochasticity of protein evolution using phage-assisted continuous evolution
- PMID: 23674678
- PMCID: PMC3670371
- DOI: 10.1073/pnas.1220670110
Experimental interrogation of the path dependence and stochasticity of protein evolution using phage-assisted continuous evolution
Abstract
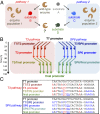
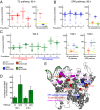
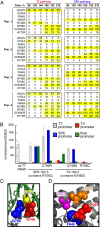
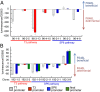
To what extent are evolutionary outcomes determined by a population's recent environment, and to what extent do they depend on historical contingency and random chance? Here we apply a unique experimental system to investigate evolutionary reproducibility and path dependence at the protein level. We combined phage-assisted continuous evolution with high-throughput sequencing to analyze evolving protein populations as they adapted to divergent and then convergent selection pressures over hundreds of generations. Independent populations of T7 RNA polymerase genes were subjected to one of two selection histories ("pathways") demanding recognition of distinct intermediate promoters followed by a common final promoter. We observed distinct classes of solutions with unequal phenotypic activity and evolutionary potential evolve from the two pathways, as well as from replicate populations exposed to identical selection conditions. Mutational analysis revealed specific epistatic interactions that explained the observed path dependence and irreproducibility. Our results reveal in molecular detail how protein adaptation to different environments, as well as stochasticity among populations evolved in the same environment, can both generate evolutionary outcomes that preclude subsequent convergence.
Keywords: directed evolution; evolutionary biology; tape of life.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
-
- Gould SJ. Wonderful Life: The Burgess Shale and the Nature of History. New York: WW Norton & Co; 1989.
-
- Travisano M, Mongold JA, Bennett AF, Lenski RE. Experimental tests of the roles of adaptation, chance, and history in evolution. Science. 1995;267(5194):87–90. - PubMed
-
- Barrick JE, et al. Genome evolution and adaptation in a long-term experiment with Escherichia coli. Nature. 2009;461(7268):1243–1247. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

