Gain-of-sensitivity mutations in a Trim5-resistant primary isolate of pathogenic SIV identify two independent conserved determinants of Trim5α specificity
- PMID: 23675300
- PMCID: PMC3649984
- DOI: 10.1371/journal.ppat.1003352
Gain-of-sensitivity mutations in a Trim5-resistant primary isolate of pathogenic SIV identify two independent conserved determinants of Trim5α specificity
Abstract
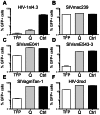
Retroviral capsid recognition by Trim5 blocks productive infection. Rhesus macaques harbor three functionally distinct Trim5 alleles: Trim5α(Q) , Trim5α(TFP) and Trim5(CypA) . Despite the high degree of amino acid identity between Trim5α(Q) and Trim5α(TFP) alleles, the Q/TFP polymorphism results in the differential restriction of some primate lentiviruses, suggesting these alleles differ in how they engage these capsids. Simian immunodeficiency virus of rhesus macaques (SIVmac) evolved to resist all three alleles. Thus, SIVmac provides a unique opportunity to study a virus in the context of the Trim5 repertoire that drove its evolution in vivo. We exploited the evolved rhesus Trim5α resistance of this capsid to identify gain-of-sensitivity mutations that distinguish targets between the Trim5α(Q) and Trim5α(TFP) alleles. While both alleles recognize the capsid surface, Trim5α(Q) and Trim5α(TFP) alleles differed in their ability to restrict a panel of capsid chimeras and single amino acid substitutions. When mapped onto the structure of the SIVmac239 capsid N-terminal domain, single amino acid substitutions affecting both alleles mapped to the β-hairpin. Given that none of the substitutions affected Trim5α(Q) alone, and the fact that the β-hairpin is conserved among retroviral capsids, we propose that the β-hairpin is a molecular pattern widely exploited by Trim5α proteins. Mutations specifically affecting rhesus Trim5α(TFP) (without affecting Trim5α(Q) ) surround a site of conservation unique to primate lentiviruses, overlapping the CPSF6 binding site. We believe targeting this site is an evolutionary innovation driven specifically by the emergence of primate lentiviruses in Africa during the last 12 million years. This modularity in targeting may be a general feature of Trim5 evolution, permitting different regions of the PRYSPRY domain to evolve independent interactions with capsid.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


Similar articles
-
TRIM5 alpha drives SIVsmm evolution in rhesus macaques.PLoS Pathog. 2013;9(8):e1003577. doi: 10.1371/journal.ppat.1003577. Epub 2013 Aug 22. PLoS Pathog. 2013. PMID: 23990789 Free PMC article.
-
Mutation in the Disordered Linker Region of Capsid Disrupts Viral Kinetics of a Neuropathogenic SIV in Rhesus Macaques.Microbiol Spectr. 2022 Apr 27;10(2):e0047822. doi: 10.1128/spectrum.00478-22. Epub 2022 Mar 17. Microbiol Spectr. 2022. PMID: 35297654 Free PMC article.
-
TRIM5alpha Modulates Immunodeficiency Virus Control in Rhesus Monkeys.PLoS Pathog. 2010 Jan 22;6(1):e1000738. doi: 10.1371/journal.ppat.1000738. PLoS Pathog. 2010. PMID: 20107597 Free PMC article.
-
Anti-retroviral activity of TRIM5 alpha.Rev Med Virol. 2010 Mar;20(2):77-92. doi: 10.1002/rmv.637. Rev Med Virol. 2010. PMID: 20049904 Review.
-
Recent insights into the mechanism and consequences of TRIM5α retroviral restriction.AIDS Res Hum Retroviruses. 2011 Mar;27(3):231-8. doi: 10.1089/AID.2010.0367. AIDS Res Hum Retroviruses. 2011. PMID: 21247355 Free PMC article. Review.
Cited by
-
Recognition of the HIV capsid by the TRIM5α restriction factor is mediated by a subset of pre-existing conformations of the TRIM5α SPRY domain.Biochemistry. 2014 Mar 11;53(9):1466-76. doi: 10.1021/bi4014962. Epub 2014 Feb 24. Biochemistry. 2014. PMID: 24506064 Free PMC article.
-
Resistance to simian immunodeficiency virus low dose rectal challenge is associated with higher constitutive TRIM5α expression in PBMC.Retrovirology. 2014 May 23;11:39. doi: 10.1186/1742-4690-11-39. Retrovirology. 2014. PMID: 24884551 Free PMC article.
-
General Model for Retroviral Capsid Pattern Recognition by TRIM5 Proteins.J Virol. 2018 Jan 30;92(4):e01563-17. doi: 10.1128/JVI.01563-17. Print 2018 Feb 15. J Virol. 2018. PMID: 29187540 Free PMC article.
-
Dynamic regulation of HIV-1 capsid interaction with the restriction factor TRIM5α identified by magic-angle spinning NMR and molecular dynamics simulations.Proc Natl Acad Sci U S A. 2018 Nov 6;115(45):11519-11524. doi: 10.1073/pnas.1800796115. Epub 2018 Oct 17. Proc Natl Acad Sci U S A. 2018. PMID: 30333189 Free PMC article.
-
Structural biology of supramolecular assemblies by magic-angle spinning NMR spectroscopy.Q Rev Biophys. 2017 Jan;50:e1. doi: 10.1017/S0033583516000159. Q Rev Biophys. 2017. PMID: 28093096 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

