Bck2 acts through the MADS box protein Mcm1 to activate cell-cycle-regulated genes in budding yeast
- PMID: 23675312
- PMCID: PMC3649975
- DOI: 10.1371/journal.pgen.1003507
Bck2 acts through the MADS box protein Mcm1 to activate cell-cycle-regulated genes in budding yeast
Abstract
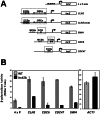
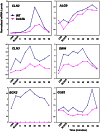
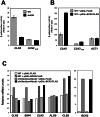
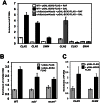
The Bck2 protein is a potent genetic regulator of cell-cycle-dependent gene expression in budding yeast. To date, most experiments have focused on assessing a potential role for Bck2 in activation of the G1/S-specific transcription factors SBF (Swi4, Swi6) and MBF (Mbp1, Swi6), yet the mechanism of gene activation by Bck2 has remained obscure. We performed a yeast two-hybrid screen using a truncated version of Bck2 and discovered six novel Bck2-binding partners including Mcm1, an essential protein that binds to and activates M/G1 promoters through Early Cell cycle Box (ECB) elements as well as to G2/M promoters. At M/G1 promoters Mcm1 is inhibited by association with two repressors, Yox1 or Yhp1, and gene activation ensues once repression is relieved by an unknown activating signal. Here, we show that Bck2 interacts physically with Mcm1 to activate genes during G1 phase. We used chromatin immunoprecipitation (ChIP) experiments to show that Bck2 localizes to the promoters of M/G1-specific genes, in a manner dependent on functional ECB elements, as well as to the promoters of G1/S and G2/M genes. The Bck2-Mcm1 interaction requires valine 69 on Mcm1, a residue known to be required for interaction with Yox1. Overexpression of BCK2 decreases Yox1 localization to the early G1-specific CLN3 promoter and rescues the lethality caused by overexpression of YOX1. Our data suggest that Yox1 and Bck2 may compete for access to the Mcm1-ECB scaffold to ensure appropriate activation of the initial suite of genes required for cell cycle commitment.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



References
-
- Laub MT, McAdams HH, Feldblyum T, Fraser CM, Shapiro L (2000) Global analysis of the genetic network controlling a bacterial cell cycle. Science 290: 2144–2148. - PubMed
-
- Cho RJ, Huang M, Campbell MJ, Dong H, Steinmetz L, et al. (2001) Transcriptional regulation and function during the human cell cycle. Nat Genet 27: 48–54. - PubMed
-
- Oliva A, Rosebrock A, Ferrezuelo F, Pyne S, Chen H, et al. (2005) The cell cycle-regulated genes of Schizosaccharomyces pombe. PLoS Biol 3: e225 doi:10.1371/journal.pbio.0030225. - DOI - PMC - PubMed
-
- Morgan DO (1997) Cyclin-dependent kinases: engines, clocks, and microprocessors. Annu Rev Cell Dev Biol 13: 261–291. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

