Genomic and proteomic analyses of the terminally redundant genome of the Pseudomonas aeruginosa phage PaP1: establishment of genus PaP1-like phages
- PMID: 23675441
- PMCID: PMC3652863
- DOI: 10.1371/journal.pone.0062933
Genomic and proteomic analyses of the terminally redundant genome of the Pseudomonas aeruginosa phage PaP1: establishment of genus PaP1-like phages
Abstract
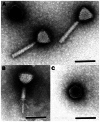
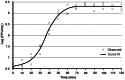
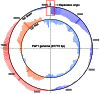
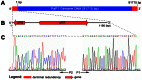
We isolated and characterized a new Pseudomonas aeruginosa myovirus named PaP1. The morphology of this phage was visualized by electron microscopy and its genome sequence and ends were determined. Finally, genomic and proteomic analyses were performed. PaP1 has an icosahedral head with an apex diameter of 68-70 nm and a contractile tail with a length of 138-140 nm. The PaP1 genome is a linear dsDNA molecule containing 91,715 base pairs (bp) with a G+C content of 49.36% and 12 tRNA genes. A strategy to identify the genome ends of PaP1 was designed. The genome has a 1190 bp terminal redundancy. PaP1 has 157 open reading frames (ORFs). Of these, 143 proteins are homologs of known proteins, but only 38 could be functionally identified. Sodium dodecyl sulfate-polyacrylamide gel electrophoresis and high-performance liquid chromatography-mass spectrometry allowed identification of 12 ORFs as structural protein coding genes within the PaP1 genome. Comparative genomic analysis indicated that the Pseudomonas aeruginosa phage PaP1, JG004, PAK_P1 and vB_PaeM_C2-10_Ab1 share great similarity. Besides their similar biological characteristics, the phages contain 123 core genes and have very close phylogenetic relationships, which distinguish them from other known phage genera. We therefore propose that these four phages be classified as PaP1-like phages, a new phage genus of Myoviridae that infects Pseudomonas aeruginosa.
Conflict of interest statement
Figures





Similar articles
-
Characterization of Two Pseudomonas aeruginosa Viruses vB_PaeM_SCUT-S1 and vB_PaeM_SCUT-S2.Viruses. 2019 Apr 1;11(4):318. doi: 10.3390/v11040318. Viruses. 2019. PMID: 30939832 Free PMC article.
-
Myoviridae bacteriophages of Pseudomonas aeruginosa: a long and complex evolutionary pathway.Res Microbiol. 2003 May;154(4):269-75. doi: 10.1016/S0923-2508(03)00070-6. Res Microbiol. 2003. PMID: 12798231
-
"phiKZ-like viruses", a proposed new genus of myovirus bacteriophages.Arch Virol. 2007;152(10):1955-9. doi: 10.1007/s00705-007-1037-7. Epub 2007 Aug 7. Arch Virol. 2007. PMID: 17680323
-
The SPO1-related bacteriophages.Arch Virol. 2010 Oct;155(10):1547-61. doi: 10.1007/s00705-010-0783-0. Epub 2010 Aug 17. Arch Virol. 2010. PMID: 20714761 Review.
-
Phage phiKZ-The First of Giants.Viruses. 2021 Jan 20;13(2):149. doi: 10.3390/v13020149. Viruses. 2021. PMID: 33498475 Free PMC article. Review.
Cited by
-
Isolation and Characterization of Lytic Pseudomonas aeruginosa Bacteriophages Isolated from Sewage Samples from Tunisia.Viruses. 2022 Oct 25;14(11):2339. doi: 10.3390/v14112339. Viruses. 2022. PMID: 36366441 Free PMC article.
-
Selection of phages and conditions for the safe phage therapy against Pseudomonas aeruginosa infections.Virol Sin. 2015 Feb;30(1):33-44. doi: 10.1007/s12250-014-3546-3. Epub 2015 Feb 5. Virol Sin. 2015. PMID: 25680443 Free PMC article.
-
Combination of genetically diverse Pseudomonas phages enhances the cocktail efficiency against bacteria.Sci Rep. 2023 Jun 1;13(1):8921. doi: 10.1038/s41598-023-36034-2. Sci Rep. 2023. PMID: 37264114 Free PMC article.
-
Transcriptomic and Metabolomics Profiling of Phage-Host Interactions between Phage PaP1 and Pseudomonas aeruginosa.Front Microbiol. 2017 Mar 30;8:548. doi: 10.3389/fmicb.2017.00548. eCollection 2017. Front Microbiol. 2017. PMID: 28421049 Free PMC article.
-
Pectobacterium Phage Jarilo Displays Broad Host Range and Represents a Novel Genus of Bacteriophages Within the Family Autographiviridae.Phage (New Rochelle). 2020 Dec 1;1(4):237-244. doi: 10.1089/phage.2020.0037. Epub 2020 Dec 16. Phage (New Rochelle). 2020. PMID: 36147289 Free PMC article.
References
-
- Lima-Mendez G, Toussaint A, Leplae R (2007) Analysis of the phage sequence space: the benefit of structured information. Virology 365: 241–249. - PubMed
-
- Hendrix RW (2003) Bacteriophage genomics. Curr Opin Microbiol 6: 506–511. - PubMed
-
- Twort A (1993) In focus, out of step: a biography of Frederick William Twort F.R.S., 1877–1950. Phoenix Mill; Dover, NH: A. Sutton. xi, 40 p.
-
- dHerelle F (1917) Sur un microbe invisible antagoniste des bacilles dysentériques. C R Acad Sci Paris 165: 373–375.
-
- Debarbieux L (2008) Experimental phage therapy in the beginning of the 21st century. Med Mal Infect 38: 421–425. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

