Protecting a transgene expression from the HAC-based vector by different chromatin insulators
- PMID: 23677492
- PMCID: PMC3771377
- DOI: 10.1007/s00018-013-1362-9
Protecting a transgene expression from the HAC-based vector by different chromatin insulators
Abstract
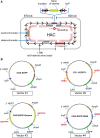
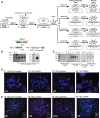
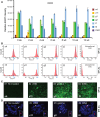
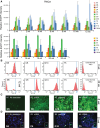
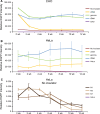
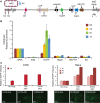
Human artificial chromosomes (HACs) are vectors that offer advantages of capacity and stability for gene delivery and expression. Several studies have even demonstrated their use for gene complementation in gene-deficient recipient cell lines and animal transgenesis. Recently, we constructed an advance HAC-based vector, alphoid(tetO)-HAC, with a conditional centromere. In this HAC, a gene-loading site was inserted into a centrochromatin domain critical for kinetochore assembly and maintenance. While by definition this domain is permissive for transcription, there have been no long-term studies on transgene expression within centrochromatin. In this study, we compared the effects of three chromatin insulators, cHS4, gamma-satellite DNA, and tDNA, on the expression of an EGFP transgene inserted into the alphoid(tetO)-HAC vector. Insulator function was essential for stable expression of the transgene in centrochromatin. In two analyzed host cell lines, a tDNA insulator composed of two functional copies of tRNA genes showed the highest barrier activity. We infer that proximity to centrochromatin does not protect genes lacking chromatin insulators from epigenetic silencing. Barrier elements that prevent gene silencing in centrochromatin would thus help to optimize transgenesis using HAC vectors.
Figures
Similar articles
-
Assembly of Multiple Full-Size Genes or Genomic DNA Fragments on Human Artificial Chromosomes Using the Iterative Integration System.Curr Protoc. 2021 Dec;1(12):e316. doi: 10.1002/cpz1.316. Curr Protoc. 2021. PMID: 34919348 Free PMC article.
-
Re-engineering an alphoid(tetO)-HAC-based vector to enable high-throughput analyses of gene function.Nucleic Acids Res. 2013 May 1;41(10):e107. doi: 10.1093/nar/gkt205. Epub 2013 Apr 4. Nucleic Acids Res. 2013. PMID: 23558748 Free PMC article.
-
Human artificial chromosome with a conditional centromere for gene delivery and gene expression.DNA Res. 2010 Oct;17(5):293-301. doi: 10.1093/dnares/dsq020. Epub 2010 Aug 26. DNA Res. 2010. PMID: 20798231 Free PMC article.
-
Human Artificial Chromosome with Regulated Centromere: A Tool for Genome and Cancer Studies.ACS Synth Biol. 2018 Sep 21;7(9):1974-1989. doi: 10.1021/acssynbio.8b00230. Epub 2018 Aug 16. ACS Synth Biol. 2018. PMID: 30075081 Free PMC article. Review.
-
HACking the centromere chromatin code: insights from human artificial chromosomes.Chromosome Res. 2012 Jul;20(5):505-19. doi: 10.1007/s10577-012-9293-0. Chromosome Res. 2012. PMID: 22825423 Review.
Cited by
-
TAR Cloning: Perspectives for Functional Genomics, Biomedicine, and Biotechnology.Mol Ther Methods Clin Dev. 2019 May 21;14:16-26. doi: 10.1016/j.omtm.2019.05.006. eCollection 2019 Sep 13. Mol Ther Methods Clin Dev. 2019. PMID: 31276008 Free PMC article. Review.
-
A pathway from chromosome transfer to engineering resulting in human and mouse artificial chromosomes for a variety of applications to bio-medical challenges.Chromosome Res. 2015 Feb;23(1):111-33. doi: 10.1007/s10577-014-9459-z. Chromosome Res. 2015. PMID: 25657031 Free PMC article. Review.
-
Fluorescence Amplification Method for Forward Genetic Discovery of Factors in Human mRNA Degradation.Mol Cell. 2017 Jan 5;65(1):191-201. doi: 10.1016/j.molcel.2016.11.032. Epub 2016 Dec 22. Mol Cell. 2017. PMID: 28017590 Free PMC article.
-
Human AlphoidtetO Artificial Chromosome as a Gene Therapy Vector for the Developing Hemophilia A Model in Mice.Cells. 2020 Apr 3;9(4):879. doi: 10.3390/cells9040879. Cells. 2020. PMID: 32260189 Free PMC article.
-
Assembly of Multiple Full-Size Genes or Genomic DNA Fragments on Human Artificial Chromosomes Using the Iterative Integration System.Curr Protoc. 2021 Dec;1(12):e316. doi: 10.1002/cpz1.316. Curr Protoc. 2021. PMID: 34919348 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

