The helicase FBH1 is tightly regulated by PCNA via CRL4(Cdt2)-mediated proteolysis in human cells
- PMID: 23677613
- PMCID: PMC3711418
- DOI: 10.1093/nar/gkt397
The helicase FBH1 is tightly regulated by PCNA via CRL4(Cdt2)-mediated proteolysis in human cells
Abstract
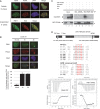
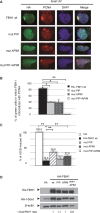
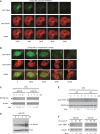
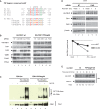
During replication, DNA damage can challenge replication fork progression and cell viability. Homologous Recombination (HR) and Translesion Synthesis (TLS) pathways appear as major players involved in the resumption and completion of DNA replication. How both pathways are coordinated in human cells to maintain genome stability is unclear. Numerous helicases are involved in HR regulation. Among them, the helicase FBH1 accumulates at sites of DNA damage and potentially constrains HR via its anti-recombinase activity. However, little is known about its regulation in vivo. Here, we report a mechanism that controls the degradation of FBH1 after DNA damage. Firstly, we found that the sliding clamp Proliferating Cell Nuclear Antigen (PCNA) is critical for FBH1 recruitment to replication factories or DNA damage sites. We then showed the anti-recombinase activity of FBH1 is partially dependent on its interaction with PCNA. Intriguingly, after its re-localization, FBH1 is targeted for degradation by the Cullin-ring ligase 4-Cdt2 (CRL4(Cdt2))-PCNA pathway via a PCNA-interacting peptide (PIP) degron. Importantly, expression of non-degradable FBH1 mutant impairs the recruitment of the TLS polymerase eta to chromatin in UV-irradiated cells. Thus, we propose that after DNA damage, FBH1 might be required to restrict HR and then degraded by the Cdt2-proteasome pathway to facilitate TLS pathway.
Figures
References
-
- Branzei D, Foiani M. RecQ helicases queuing with Srs2 to disrupt Rad51 filaments and suppress recombination. Genes Dev. 2007;21:3019–3026. - PubMed
-
- Krejci L, Van Komen S, Li Y, Villemain J, Reddy MS, Klein H, Ellenberger T, Sung P. DNA helicase Srs2 disrupts the Rad51 presynaptic filament. Nature. 2003;423:305–309. - PubMed
-
- Veaute X, Jeusset J, Soustelle C, Kowalczykowski SC, Le Cam E, Fabre F. The Srs2 helicase prevents recombination by disrupting Rad51 nucleoprotein filaments. Nature. 2003;423:309–312. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

