Regulation of CCN1 via the 3'-untranslated region
- PMID: 23677691
- PMCID: PMC3709046
- DOI: 10.1007/s12079-013-0202-x
Regulation of CCN1 via the 3'-untranslated region
Abstract
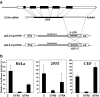
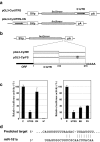
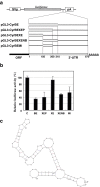
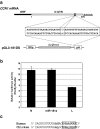
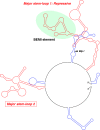
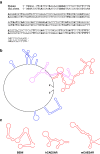
The 3'-untranslated region (UTR) is known to be a critical regulator of post-transcriptional events that determine the gene expression at the RNA level. The gene CCN1 is one of the classical members of the matricellular CCN family and is involved in a number of biological processes during mammalian development. In the present study, the 600-bp 3'-UTR of CCN1 was functionally characterized. Reporter gene analysis revealed that the entire 3'-UTR profoundly repressed gene expression in cis in different types of the cells, to which both the proximal and distal-halves of the 3'-UTR segments contributed almost equally. Deletion analysis of the 3'-UTR indicated a distinct functional element in the proximal half, whereas a putative target for microRNA-181s was predicted in silico in the distal half. Of note, the repressive RNA element in the proximal half was shown to be capable of forming a stable secondary structure. However, unexpectedly, a reporter construct with a tandem repeat of the predicted miR-181 targets failed to respond to miR-181a. In addition, the other major structured element predicted in the distal half was similarly characterized. To our surprise, the second element rather enhanced the reporter gene expression in cis. These results indicate the involvement of multiple regulatory elements in the CCN1 3'-UTR and suggest the complexity of the miRNA action as well as the 3'-UTR-mediated gene regulation.
Figures

Similar articles
-
Single and Compound Knock-outs of MicroRNA (miRNA)-155 and Its Angiogenic Gene Target CCN1 in Mice Alter Vascular and Neovascular Growth in the Retina via Resident Microglia.J Biol Chem. 2015 Sep 18;290(38):23264-81. doi: 10.1074/jbc.M115.646950. Epub 2015 Aug 4. J Biol Chem. 2015. PMID: 26242736 Free PMC article.
-
Identification and characterization of cis-acting elements in the human and bovine PTH mRNA 3'-untranslated region.J Bone Miner Res. 2005 May;20(5):858-66. doi: 10.1359/JBMR.041227. Epub 2004 Dec 20. J Bone Miner Res. 2005. PMID: 15824859
-
Identification of an RNA element that confers post-transcriptional repression of connective tissue growth factor/hypertrophic chondrocyte specific 24 (ctgf/hcs24) gene: similarities to retroviral RNA-protein interactions.Oncogene. 2000 Sep 28;19(41):4773-86. doi: 10.1038/sj.onc.1203835. Oncogene. 2000. PMID: 11032028
-
Cis-acting elements in its 3' UTR mediate post-transcriptional regulation of KRAS.Oncotarget. 2016 Mar 15;7(11):11770-84. doi: 10.18632/oncotarget.7599. Oncotarget. 2016. PMID: 26930719 Free PMC article.
-
Molecular characterization of a novel androgen receptor transgene responsive to MicroRNA mediated post-transcriptional control exerted via 3'-untranslated region.Prostate. 2016 Jun;76(9):834-44. doi: 10.1002/pros.23174. Epub 2016 Mar 14. Prostate. 2016. PMID: 26988939
Cited by
-
Single and Compound Knock-outs of MicroRNA (miRNA)-155 and Its Angiogenic Gene Target CCN1 in Mice Alter Vascular and Neovascular Growth in the Retina via Resident Microglia.J Biol Chem. 2015 Sep 18;290(38):23264-81. doi: 10.1074/jbc.M115.646950. Epub 2015 Aug 4. J Biol Chem. 2015. PMID: 26242736 Free PMC article.
-
G-quadruplexes Stabilization Upregulates CCN1 and Accelerates Aging in Cultured Cerebral Endothelial Cells.Front Aging. 2022 Jan 12;2:797562. doi: 10.3389/fragi.2021.797562. eCollection 2021. Front Aging. 2022. PMID: 35822045 Free PMC article.
-
Analyses of the Posttranscriptional Regulation of CCN Genes: Approach to Multiple Steps of CCN2 Gene Expression.Methods Mol Biol. 2023;2582:127-155. doi: 10.1007/978-1-0716-2744-0_10. Methods Mol Biol. 2023. PMID: 36370348
References
LinkOut - more resources
Full Text Sources
Other Literature Sources

