Proposals for the classification of human rhinovirus species A, B and C into genotypically assigned types
- PMID: 23677786
- PMCID: PMC3749525
- DOI: 10.1099/vir.0.053686-0
Proposals for the classification of human rhinovirus species A, B and C into genotypically assigned types
Abstract
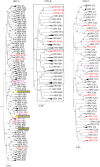
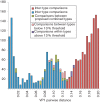
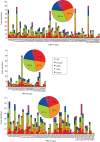
Human rhinoviruses (HRVs) frequently cause mild upper respiratory tract infections and more severe disease manifestations such as bronchiolitis and asthma exacerbations. HRV is classified into three species within the genus Enterovirus of the family Picornaviridae. HRV species A and B contain 75 and 25 serotypes identified by cross-neutralization assays, although the use of such assays for routine HRV typing is hampered by the large number of serotypes, replacement of virus isolation by molecular methods in HRV diagnosis and the poor or absent replication of HRV species C in cell culture. To address these problems, we propose an alternative, genotypic classification of HRV-based genetic relatedness analogous to that used for enteroviruses. Nucleotide distances between 384 complete VP1 sequences of currently assigned HRV (sero)types identified divergence thresholds of 13, 12 and 13 % for species A, B and C, respectively, that divided inter- and intra-type comparisons. These were paralleled by 10, 9.5 and 10 % thresholds in the larger dataset of >3800 VP4 region sequences. Assignments based on VP1 sequences led to minor revisions of existing type designations (such as the reclassification of serotype pairs, e.g. A8/A95 and A29/A44, as single serotypes) and the designation of new HRV types A101-106, B101-103 and C34-C51. A protocol for assignment and numbering of new HRV types using VP1 sequences and the restriction of VP4 sequence comparisons to type identification and provisional type assignments is proposed. Genotypic assignment and identification of HRV types will be of considerable value in the future investigation of type-associated differences in disease outcomes, transmission and epidemiology.
Figures




References
-
- Brown B. A., Maher K., Flemister M. R., Naraghi-Arani P., Uddin M., Oberste M. S., Pallansch M. A. (2009). Resolving ambiguities in genetic typing of human enterovirus species C clinical isolates and identification of enterovirus 96, 99 and 102. J Gen Virol 90, 1713–1723 10.1099/vir.0.008540-0 - DOI - PubMed
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

