Translation regulation gets its 'omics' moment
- PMID: 23677826
- PMCID: PMC3797170
- DOI: 10.1002/wrna.1173
Translation regulation gets its 'omics' moment
Abstract
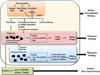
The fate of cellular RNA is largely determined by complex networks of protein-RNA interactions through ribonucleoprotein (RNP) complexes. Despite their relatively short half-life, transcripts associate with many different proteins that process, modify, translate, and degrade the RNA. Following biogenesis some mRNPs are immediately directed to translation and produce proteins, but many are diverted and regulated by processes including miRNA-mediated mechanisms, transport and localization, as well as turnover. Because of this complex interplay estimates of steady-state expression by methods such as RNAseq alone cannot capture critical aspects of cellular fate, environmental response, tumorigenesis, or gene expression regulation. More selective and integrative tools are needed to measure protein-RNA complexes and the regulatory processes involved. One focus area is measurements of the transcriptome associated with ribosomes and translation. These so-called polysome or ribosome profiling techniques can evaluate translation efficiency as well as the interplay between translation initiation, elongation, and termination-subject areas not well understood at a systems biology level. Ribosome profiling is a highly promising technique that provides mRNA positional information of ribosome occupancy, potentially bridging the gap between gene expression (i.e., RNAseq and microarray analysis) and protein quantification (i.e., mass spectrometry). In combination with methods such as RNA immunoprecipitation, miRNA profiling, or proteomics, we obtain a fresh view of global post-transcriptional and translational gene regulation. In addition, these techniques also provide new insight into new regulatory elements, such as alternative open reading frames, and translation regulation under different conditions.
Copyright © 2013 John Wiley & Sons, Ltd.
Figures

References
-
- Costa FF. Non-coding RNAs: Meet thy masters. Bioessays. 2010 Jul;32(7):599–608. Review. - PubMed
-
- Castello A, Fischer B, Hentze MW, Preiss T. RNA-binding proteins in Mendelian disease. Trends Genet. 2013 Feb 14; - PubMed
-
- Melamed D, Arava Y. Genome-wide analysis of mRNA polysomal profiles with spotted DNA microarrays. Methods Enzymol. 2007;431:177–201. Review. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

