HIV-2 and SIVmac accessory virulence factor Vpx down-regulates SAMHD1 enzyme catalysis prior to proteasome-dependent degradation
- PMID: 23677995
- PMCID: PMC3696684
- DOI: 10.1074/jbc.M113.469007
HIV-2 and SIVmac accessory virulence factor Vpx down-regulates SAMHD1 enzyme catalysis prior to proteasome-dependent degradation
Abstract
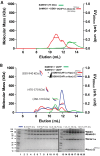
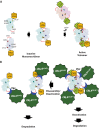
SAMHD1, a dGTP-regulated deoxyribonucleoside triphosphate (dNTP) triphosphohydrolase, down-regulates dNTP pools in terminally differentiated and quiescent cells, thereby inhibiting HIV-1 infection at the reverse transcription step. HIV-2 and simian immunodeficiency virus (SIV) counteract this restriction via a virion-associated virulence accessory factor, Vpx (Vpr in some SIVs), which loads SAMHD1 onto CRL4-DCAF1 E3 ubiquitin ligase for polyubiquitination, programming it for proteasome-dependent degradation. However, the detailed molecular mechanisms of SAMHD1 recruitment to the E3 ligase have not been defined. Further, whether divergent, orthologous Vpx proteins, encoded by distinct HIV/SIV strains, bind SAMHD1 in a similar manner, at a molecular level, is not known. We applied surface plasmon resonance analysis to assess the requirements for and kinetics of binding between various primate SAMHD1 proteins and Vpx proteins from SIV or HIV-2 strains. Our data indicate that Vpx proteins, bound to DCAF1, interface with the C terminus of primate SAMHD1 proteins with nanomolar affinity, manifested by rapid association and slow dissociation. Further, we provide evidence that Vpx binding to SAMHD1 inhibits its catalytic activity and induces disassembly of a dGTP-dependent oligomer. Our studies reveal a previously unrecognized biochemical mechanism of Vpx-mediated SAMHD1 inhibition: direct down-modulation of its catalytic activity, mediated by the same binding event that leads to SAMHD1 recruitment to the E3 ubiquitin ligase for proteasome-dependent degradation.
Keywords: Enzyme Turnover; Enzymes; HIV; Restriction Factors; Ubiquitin Ligase; Ubiquitination; Virulence Factors.
Figures

Similar articles
-
HIV/simian immunodeficiency virus (SIV) accessory virulence factor Vpx loads the host cell restriction factor SAMHD1 onto the E3 ubiquitin ligase complex CRL4DCAF1.J Biol Chem. 2012 Apr 6;287(15):12550-8. doi: 10.1074/jbc.M112.340711. Epub 2012 Feb 23. J Biol Chem. 2012. PMID: 22362772 Free PMC article.
-
Inhibition of Vpx-Mediated SAMHD1 and Vpr-Mediated Host Helicase Transcription Factor Degradation by Selective Disruption of Viral CRL4 (DCAF1) E3 Ubiquitin Ligase Assembly.J Virol. 2017 Apr 13;91(9):e00225-17. doi: 10.1128/JVI.00225-17. Print 2017 May 1. J Virol. 2017. PMID: 28202763 Free PMC article.
-
Evolutionary toggling of Vpx/Vpr specificity results in divergent recognition of the restriction factor SAMHD1.PLoS Pathog. 2013;9(7):e1003496. doi: 10.1371/journal.ppat.1003496. Epub 2013 Jul 18. PLoS Pathog. 2013. PMID: 23874202 Free PMC article.
-
Lentivirus Vpr and Vpx accessory proteins usurp the cullin4-DDB1 (DCAF1) E3 ubiquitin ligase.Curr Opin Virol. 2012 Dec;2(6):755-63. doi: 10.1016/j.coviro.2012.09.010. Epub 2012 Oct 10. Curr Opin Virol. 2012. PMID: 23062609 Free PMC article. Review.
-
[Research Progress in Viral Protein Vpx induction of Proteasomal Degradation of the Antiviral Factor SAMHD1].Bing Du Xue Bao. 2016 May;32(3):355-60. Bing Du Xue Bao. 2016. PMID: 29963825 Review. Chinese.
Cited by
-
The aryl hydrocarbon receptor and interferon gamma generate antiviral states via transcriptional repression.Elife. 2018 Aug 22;7:e38867. doi: 10.7554/eLife.38867. Elife. 2018. PMID: 30132758 Free PMC article.
-
New insights into an X-traordinary viral protein.Front Microbiol. 2014 Apr 8;5:126. doi: 10.3389/fmicb.2014.00126. eCollection 2014. Front Microbiol. 2014. PMID: 24782834 Free PMC article. Review.
-
SLX4-SLX1 Protein-independent Down-regulation of MUS81-EME1 Protein by HIV-1 Viral Protein R (Vpr).J Biol Chem. 2016 Aug 12;291(33):16936-16947. doi: 10.1074/jbc.M116.721183. Epub 2016 Jun 27. J Biol Chem. 2016. PMID: 27354282 Free PMC article.
-
SAMHD1: Recurring roles in cell cycle, viral restriction, cancer, and innate immunity.Autoimmunity. 2018 May;51(3):96-110. doi: 10.1080/08916934.2018.1454912. Epub 2018 Mar 27. Autoimmunity. 2018. PMID: 29583030 Free PMC article. Review.
-
Cellular and Biochemical Mechanisms of the Retroviral Restriction Factor SAMHD1.ISRN Biochem. 2013 Jul 7;2013:728392. doi: 10.1155/2013/728392. ISRN Biochem. 2013. PMID: 24523981 Free PMC article.
References
-
- Berger A., Sommer A. F., Zwarg J., Hamdorf M., Welzel K., Esly N., Panitz S., Reuter A., Ramos I., Jatiani A., Mulder L. C., Fernandez-Sesma A., Rutsch F., Simon V., König R., Flory E. (2011) SAMHD1-deficient CD14+ cells from individuals with Aicardi-Goutieres syndrome are highly susceptible to HIV-1 infection. PLoS Pathog. 7, e1002425. - PMC - PubMed
-
- Baldauf H. M., Pan X., Erikson E., Schmidt S., Daddacha W., Burggraf M., Schenkova K., Ambiel I., Wabnitz G., Gramberg T., Panitz S., Flory E., Landau N. R., Sertel S., Rutsch F., Lasitschka F., Kim B., König R., Fackler O. T., Keppler O. T. (2012) SAMHD1 restricts HIV-1 infection in resting CD4+ T cells. Nat. Med. 18, 1682–1687 - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

