Evolutionary changes in chlorophyllide a oxygenase (CAO) structure contribute to the acquisition of a new light-harvesting complex in micromonas
- PMID: 23677999
- PMCID: PMC3707637
- DOI: 10.1074/jbc.M113.462663
Evolutionary changes in chlorophyllide a oxygenase (CAO) structure contribute to the acquisition of a new light-harvesting complex in micromonas
Abstract
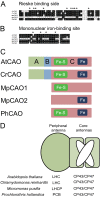
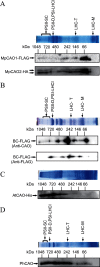
Chlorophyll b is found in photosynthetic prokaryotes and primary and secondary endosymbionts, although their light-harvesting systems are quite different. Chlorophyll b is synthesized from chlorophyll a by chlorophyllide a oxygenase (CAO), which is a Rieske-mononuclear iron oxygenase. Comparison of the amino acid sequences of CAO among photosynthetic organisms elucidated changes in the domain structures of CAO during evolution. However, the evolutionary relationship between the light-harvesting system and the domain structure of CAO remains unclear. To elucidate this relationship, we investigated the CAO structure and the pigment composition of chlorophyll-protein complexes in the prasinophyte Micromonas. The Micromonas CAO is composed of two genes, MpCAO1 and MpCAO2, that possess Rieske and mononuclear iron-binding motifs, respectively. Only when both genes were introduced into the chlorophyll b-less Arabidopsis mutant (ch1-1) was chlorophyll b accumulated, indicating that cooperation between the two subunits is required to synthesize chlorophyll b. Although Micromonas has a characteristic light-harvesting system in which chlorophyll b is incorporated into the core antennas of reaction centers, chlorophyll b was also incorporated into the core antennas of reaction centers of the Arabidopsis transformants that contained the two Micromonas CAO proteins. Based on these results, we discuss the evolutionary relationship between the structures of CAO and light-harvesting systems.
Keywords: CAO; Chlorophyll; Chloroplast; Enzymes; Evolution; Photosynthesis; Photosynthetic Pigments; Photosystem.
Figures





Similar articles
-
Structural Characterization of the Chlorophyllide a Oxygenase (CAO) Enzyme Through an In Silico Approach.J Mol Evol. 2023 Apr;91(2):225-235. doi: 10.1007/s00239-023-10100-9. Epub 2023 Mar 3. J Mol Evol. 2023. PMID: 36869271
-
Functional analysis of N-terminal domains of Arabidopsis chlorophyllide a oxygenase.Plant Physiol Biochem. 2007 Oct-Nov;45(10-11):740-9. doi: 10.1016/j.plaphy.2007.07.016. Epub 2007 Jul 29. Plant Physiol Biochem. 2007. PMID: 17884554
-
Compensation Mechanism of the Photosynthetic Apparatus in Arabidopsis thaliana ch1 Mutants.Int J Mol Sci. 2020 Dec 28;22(1):221. doi: 10.3390/ijms22010221. Int J Mol Sci. 2020. PMID: 33379339 Free PMC article.
-
Chlorophyll cycle regulates the construction and destruction of the light-harvesting complexes.Biochim Biophys Acta. 2011 Aug;1807(8):968-76. doi: 10.1016/j.bbabio.2011.01.002. Epub 2011 Jan 7. Biochim Biophys Acta. 2011. PMID: 21216224 Review.
-
Photosynthetic acclimation: structural reorganisation of light harvesting antenna--role of redox-dependent phosphorylation of major and minor chlorophyll a/b binding proteins.FEBS J. 2008 Mar;275(6):1056-68. doi: 10.1111/j.1742-4658.2008.06262.x. FEBS J. 2008. PMID: 18318833 Review.
Cited by
-
Brchli1 mutation induces bright yellow leaves by disrupting magnesium chelatase I subunit function in Chinese cabbage (Brassica rapa L. ssp. pekinensis).Front Plant Sci. 2024 Aug 30;15:1450242. doi: 10.3389/fpls.2024.1450242. eCollection 2024. Front Plant Sci. 2024. PMID: 39280951 Free PMC article.
-
Identification of a biomass unaffected pale green mutant gene in Chinese cabbage (Brassica rapa L. ssp. pekinensis).Sci Rep. 2022 May 11;12(1):7731. doi: 10.1038/s41598-022-11825-1. Sci Rep. 2022. PMID: 35546169 Free PMC article.
-
Heterologous synthesis of chlorophyll b in Nannochloropsis salina enhances growth and lipid production by increasing photosynthetic efficiency.Biotechnol Biofuels. 2019 May 14;12:122. doi: 10.1186/s13068-019-1462-3. eCollection 2019. Biotechnol Biofuels. 2019. PMID: 31114631 Free PMC article.
-
Crystal Structure and Catalytic Mechanism of 7-Hydroxymethyl Chlorophyll a Reductase.J Biol Chem. 2016 Jun 17;291(25):13349-59. doi: 10.1074/jbc.M116.720342. Epub 2016 Apr 12. J Biol Chem. 2016. PMID: 27072131 Free PMC article.
-
Responses of unicellular predators to cope with the phototoxicity of photosynthetic prey.Nat Commun. 2019 Dec 6;10(1):5606. doi: 10.1038/s41467-019-13568-6. Nat Commun. 2019. PMID: 31811209 Free PMC article.
References
-
- Neilson J. A., Durnford D. G. (2010) Structural and functional diversification of the light-harvesting complexes in photosynthetic eukaryotes. Photosynth. Res. 106, 57–71 - PubMed
-
- Green B. R., Durnford D. G. (1996) The chlorophyll-carotenoid proteins of oxygenic photosynthesis. Annu. Rev. Plant Physiol. Plant Mol. Biol. 47, 685–714 - PubMed
-
- Renger T., Schlodder E. (2010) Primary photophysical processes in photosystem II: bridging the gap between crystal structure and optical spectra. Chemphyschem 11, 1141–1153 - PubMed
-
- Chen M., Schliep M., Willows R. D., Cai Z. L., Neilan B. A., Scheer H. (2010) A red-shifted chlorophyll. Science 329, 1318–1319 - PubMed
-
- Adir N. (2005) Elucidation of the molecular structures of components of the phycobilisome: reconstructing a giant. Photosynth. Res. 85, 15–32 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

