Epstein-Barr virus-encoded microRNA BART15-3p promotes cell apoptosis partially by targeting BRUCE
- PMID: 23678170
- PMCID: PMC3700184
- DOI: 10.1128/JVI.03159-12
Epstein-Barr virus-encoded microRNA BART15-3p promotes cell apoptosis partially by targeting BRUCE
Abstract
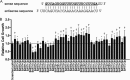
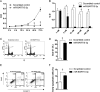
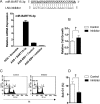
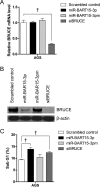
Epstein-Barr Virus (EBV) generates a variety of viral microRNAs (miRNAs) by processing the BHRF1 and BamHI A rightward (BART) transcripts. BART miRNAs are expressed in all cells latently infected with EBV, but the functions of most BART miRNAs remain unknown. The results of a cell proliferation assay revealed that miR-BART15-3p inhibited cell proliferation. Fluorescence-activated cell sorting following staining with annexin V or propidium iodide showed that miR-BART15-3p promoted apoptosis. Furthermore, the inhibitor for miR-BART15-3p increased cell growth and reduced apoptosis in EBV-infected cells. Using bioinformatic analyses, we predicted that miR-BART15-3p may target the antiapoptotic B-cell lymphoma 2 (BCL2), BCL2L2, DEAD (Asp-Glu-Ala-Asp) box polypeptide 42 (DDX42), and baculovirus inhibitor of apoptosis repeat-containing ubiquitin-conjugating enzyme (BRUCE) mRNAs. The luciferase reporter assay showed that only the 3' untranslated region (UTR) of BRUCE was affected by miR-BART15-3p. Two putative seed-matched sites for miR-BART15-3p were evident on the BRUCE 3' UTR. The results of a mutation study indicated that miR-BART15-3p hybridized only with the first seed-matched site on the BRUCE 3' UTR. miR-BART15-3p downregulated the BRUCE protein in EBV-negative cells, while the inhibitor for miR-BART15-3p upregulated the BRUCE protein in EBV-infected cells without affecting the BRUCE mRNA level. miR-BART15-3p was secreted from EBV-infected gastric carcinoma cells, and the level of miR-BART15-3p was 2- to 16-fold higher in exosomes than in the corresponding cells. Our data suggest that miR-BART15-3p can induce apoptosis partially by inhibiting the translation of the apoptosis inhibitor BRUCE. Further study is warranted to understand the role of miR-BART15-3p in the EBV life cycle.
Figures




References
-
- Bartel DP. 2004. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116:281–297 - PubMed
-
- Young LS, Rickinson AB. 2004. Epstein-Barr virus: 40 years on. Nat. Rev. Cancer 4:757–768 - PubMed
-
- Young LS, Murray PG. 2003. Epstein-Barr virus and oncogenesis: from latent genes to tumours. Oncogene 22:5108–5121 - PubMed
-
- Middeldorp JM, Brink AA, van den Brule AJ, Meijer CJ. 2003. Pathogenic roles for Epstein-Barr virus (EBV) gene products in EBV-associated proliferative disorders. Crit. Rev. Oncol. Hematol. 45:1–36 - PubMed
-
- van Beek J, Brink AA, Vervoort MB, van Zijp MJ, Meijer CJ, van den Brule AJ, Middeldorp JM. 2003. In vivo transcription of the Epstein-Barr virus (EBV) BamHI-A region without associated in vivo BARF0 protein expression in multiple EBV-associated disorders. J. Gen. Virol. 84:2647–2659 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

