Ligand-dependent activation and deactivation of the human adenosine A(2A) receptor
- PMID: 23678995
- PMCID: PMC4120839
- DOI: 10.1021/ja404391q
Ligand-dependent activation and deactivation of the human adenosine A(2A) receptor
Abstract
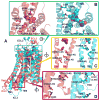
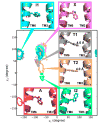
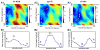
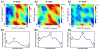
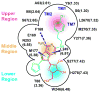
G-protein-coupled receptors (GPCRs) are membrane proteins with critical functions in cellular signal transduction, representing a primary class of drug targets. Acting by direct binding, many drugs modulate GPCR activity and influence the signaling pathways associated with numerous diseases. However, complete details of ligand-dependent GPCR activation/deactivation are difficult to obtain from experiments. Therefore, it remains unclear how ligands modulate a GPCR's activity. To elucidate the ligand-dependent activation/deactivation mechanism of the human adenosine A2A receptor (AA2AR), a member of the class A GPCRs, we performed large-scale unbiased molecular dynamics and metadynamics simulations of the receptor embedded in a membrane. At the atomic level, we have observed distinct structural states that resemble the active and inactive states. In particular, we noted key structural elements changing in a highly concerted fashion during the conformational transitions, including six conformational states of a tryptophan (Trp246(6.48)). Our findings agree with a previously proposed view that, during activation, this tryptophan residue undergoes a rotameric transition that may be coupled to a series of coherent conformational changes, resulting in the opening of the G-protein binding site. Further, metadynamics simulations provide quantitative evidence for this mechanism, suggesting how ligand binding shifts the equilibrium between the active and inactive states. Our analysis also proposes that a few specific residues are associated with agonism/antagonism, affinity, and selectivity, and suggests that the ligand-binding pocket can be thought of as having three distinct regions, providing dynamic features for structure-based design. Additional simulations with AA2AR bound to a novel ligand are consistent with our proposed mechanism. Generally, our study provides insights into the ligand-dependent AA2AR activation/deactivation in addition to what has been found in crystal structures. These results should aid in the discovery of more effective and selective GPCR ligands.
Figures


Similar articles
-
Metadynamics simulations leveraged by statistical analyses and artificial intelligence-based tools to inform the discovery of G protein-coupled receptor ligands.Front Endocrinol (Lausanne). 2022 Dec 23;13:1099715. doi: 10.3389/fendo.2022.1099715. eCollection 2022. Front Endocrinol (Lausanne). 2022. PMID: 36619585 Free PMC article. Review.
-
Molecular dynamics simulations reveal insights into key structural elements of adenosine receptors.Biochemistry. 2011 May 17;50(19):4194-208. doi: 10.1021/bi200100t. Epub 2011 Apr 20. Biochemistry. 2011. PMID: 21480628
-
GPCR 3D homology models for ligand screening: lessons learned from blind predictions of adenosine A2a receptor complex.Proteins. 2010 Jan;78(1):197-211. doi: 10.1002/prot.22507. Proteins. 2010. PMID: 20063437 Free PMC article.
-
Integrating Pharmacophore into Membrane Molecular Dynamics Simulations to Improve Homology Modeling of G Protein-coupled Receptors with Ligand Selectivity: A2A Adenosine Receptor as an Example.Chem Biol Drug Des. 2015 Dec;86(6):1438-50. doi: 10.1111/cbdd.12607. Epub 2015 Jul 14. Chem Biol Drug Des. 2015. PMID: 26072970
-
Molecular Biophysics of Class A G Protein Coupled Receptors-Lipids Interactome at a Glance-Highlights from the A2A Adenosine Receptor.Biomolecules. 2023 Jun 7;13(6):957. doi: 10.3390/biom13060957. Biomolecules. 2023. PMID: 37371538 Free PMC article. Review.
Cited by
-
Communication over the network of binary switches regulates the activation of A2A adenosine receptor.PLoS Comput Biol. 2015 Feb 9;11(2):e1004044. doi: 10.1371/journal.pcbi.1004044. eCollection 2015 Feb. PLoS Comput Biol. 2015. PMID: 25664580 Free PMC article.
-
Agonist Binding and G Protein Coupling in Histamine H2 Receptor: A Molecular Dynamics Study.Int J Mol Sci. 2020 Sep 12;21(18):6693. doi: 10.3390/ijms21186693. Int J Mol Sci. 2020. PMID: 32932742 Free PMC article.
-
Reconstruction of apo A2A receptor activation pathways reveal ligand-competent intermediates and state-dependent cholesterol hotspots.Sci Rep. 2019 Oct 2;9(1):14199. doi: 10.1038/s41598-019-50752-6. Sci Rep. 2019. PMID: 31578448 Free PMC article.
-
Metadynamics simulations leveraged by statistical analyses and artificial intelligence-based tools to inform the discovery of G protein-coupled receptor ligands.Front Endocrinol (Lausanne). 2022 Dec 23;13:1099715. doi: 10.3389/fendo.2022.1099715. eCollection 2022. Front Endocrinol (Lausanne). 2022. PMID: 36619585 Free PMC article. Review.
-
Deciphering the Allosteric Effect of Antagonist Vismodegib on Smoothened Receptor Deactivation Using Metadynamics Simulation.Front Chem. 2019 Jun 4;7:406. doi: 10.3389/fchem.2019.00406. eCollection 2019. Front Chem. 2019. PMID: 31214579 Free PMC article.
References
-
- Vauquelin G, von Mentzer B. G protein-coupled receptors: molecular pharmacology from academic concept to pharmaceutical research. John Wiley & Sons; 2007.
-
- Dorsam RT, Gutkind JS. Nat Rev Cancer. 2007;7:79. - PubMed
-
- Thathiah A, De Strooper B. Nat Rev Neurosci. 2011;12:73. - PubMed
-
- Vassart G, Costagliola S. Nat Rev Endocrinol. 2011;7:362. - PubMed
-
- Rask-Andersen M, Almen MS, Schioth HB. Nat Rev Drug Discov. 2011;10:579. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

