Disk-based k-mer counting on a PC
- PMID: 23679007
- PMCID: PMC3680041
- DOI: 10.1186/1471-2105-14-160
Disk-based k-mer counting on a PC
Abstract
Background: The k-mer counting problem, which is to build the histogram of occurrences of every k-symbol long substring in a given text, is important for many bioinformatics applications. They include developing de Bruijn graph genome assemblers, fast multiple sequence alignment and repeat detection.
Results: We propose a simple, yet efficient, parallel disk-based algorithm for counting k-mers. Experiments show that it usually offers the fastest solution to the considered problem, while demanding a relatively small amount of memory. In particular, it is capable of counting the statistics for short-read human genome data, in input gzipped FASTQ file, in less than 40 minutes on a PC with 16 GB of RAM and 6 CPU cores, and for long-read human genome data in less than 70 minutes. On a more powerful machine, using 32 GB of RAM and 32 CPU cores, the tasks are accomplished in less than half the time. No other algorithm for most tested settings of this problem and mammalian-size data can accomplish this task in comparable time. Our solution also belongs to memory-frugal ones; most competitive algorithms cannot efficiently work on a PC with 16 GB of memory for such massive data.
Conclusions: By making use of cheap disk space and exploiting CPU and I/O parallelism we propose a very competitive k-mer counting procedure, called KMC. Our results suggest that judicious resource management may allow to solve at least some bioinformatics problems with massive data on a commodity personal computer.
Figures
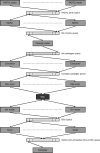





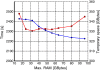
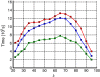
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

