Quantitative and Sensitive Detection of Cancer Genome Amplifications from Formalin Fixed Paraffin Embedded Tumors with Droplet Digital PCR
- PMID: 23682346
- PMCID: PMC3653435
- DOI: 10.4172/2161-1025.1000107
Quantitative and Sensitive Detection of Cancer Genome Amplifications from Formalin Fixed Paraffin Embedded Tumors with Droplet Digital PCR
Abstract
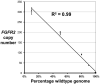
For the analysis of cancer, there is great interest in rapid and accurate detection of cancer genome amplifications containing oncogenes that are potential therapeutic targets. The vast majority of cancer tissue samples are formalin fixed and paraffin embedded (FFPE) which enables histopathological examination and long term archiving. However, FFPE cancer genomic DNA is oftentimes degraded and generally a poor substrate for many molecular biology assays. To overcome the issues of poor DNA quality from FFPE samples and detect oncogenic copy number amplifications with high accuracy and sensitivity, we developed a novel approach. Our assay requires nanogram amounts of genomic DNA, thus facilitating study of small amounts of clinical samples. Using droplet digital PCR (ddPCR), we can determine the relative copy number of specific genomic loci even in the presence of intermingled normal tissue. We used a control dilution series to determine the limits of detection for the ddPCR assay and report its improved sensitivity on minimal amounts of DNA compared to standard real-time PCR. To develop this approach, we designed an assay for the fibroblast growth factor receptor 2 gene (FGFR2) that is amplified in a gastric and breast cancers as well as others. We successfully utilized ddPCR to ascertain FGFR2 amplifications from FFPE-preserved gastrointestinal adenocarcinomas.
Keywords: archival cancer samples; cancer; copy number variations; gene amplifications.
Figures


References
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
