3' cap-independent translation enhancers of plant viruses
- PMID: 23682606
- PMCID: PMC4034384
- DOI: 10.1146/annurev-micro-092412-155609
3' cap-independent translation enhancers of plant viruses
Abstract
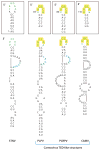
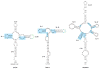
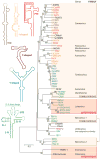
In the absence of a 5' cap, plant positive-strand RNA viruses have evolved a number of different elements in their 3' untranslated region (UTR) to attract initiation factors and/or ribosomes to their templates. These 3' cap-independent translational enhancers (3' CITEs) take different forms, such as I-shaped, Y-shaped, T-shaped, or pseudoknotted structures, or radiate multiple helices from a central hub. Common features of most 3' CITEs include the ability to bind a component of the translation initiation factor eIF4F complex and to engage in an RNA-RNA kissing-loop interaction with a hairpin loop located at the 5' end of the RNA. The two T-shaped structures can bind to ribosomes and ribosomal subunits, with one structure also able to engage in a simultaneous long-distance RNA-RNA interaction. Several of these 3' CITEs are interchangeable and there is evidence that natural recombination allows exchange of modular CITE units, which may overcome genetic resistance or extend the virus's host range.
Figures


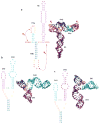
References
-
- Batten JS, Desvoyes B, Yamamura Y, Scholthof KB. A translational enhancer element on the 3 ′-proximal end of the Panicum mosaic virus genome. FEBS Lett. 2006;580:2591–97. - PubMed
-
- Diaz JA, Nieto C, Moriones E, Truniger V, Aranda MA. Molecular characterization of a Melon necrotic spot virus strain that overcomes the resistance in melon and nonhost plants. Mol Plant-Microbe Interact. 2004;17:668–75. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

