Human GGT2 does not autocleave into a functional enzyme: A cautionary tale for interpretation of microarray data on redox signaling
- PMID: 23682772
- PMCID: PMC3852618
- DOI: 10.1089/ars.2012.4997
Human GGT2 does not autocleave into a functional enzyme: A cautionary tale for interpretation of microarray data on redox signaling
Abstract
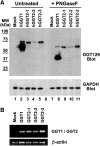
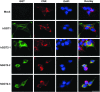
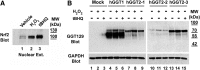
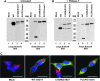
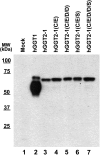
Aims: Human γ-glutamyltranspeptidase 1 (hGGT1) is a cell-surface enzyme that is a regulator of redox adaptation and drug resistance due to its glutathionase activity. The human GGT2 gene encodes a protein that is 94% identical to the amino-acid sequence of hGGT1. Transcriptional profiling analyses in a series of recent publications have implicated the hGGT2 enzyme as a modulator of disease processes. However, hGGT2 has never been shown to encode a protein with enzymatic activity. The aim of this study was to express the protein encoded by hGGT2 and each of its known variants and to assess their stability, cellular localization, and enzymatic activity.
Results: We discovered that the proteins encoded by hGGT2 and its variants are inactive propeptides. We show that hGGT2 cDNAs are transcribed with a similar efficiency to hGGT1, and the expressed propeptides are N-glycosylated. However, they do not autocleave into heterodimers, fail to localize to the plasma membrane, and do not metabolize γ-glutamyl substrates. Substituting the coding sequence of hGGT1 to conform to alterations in a CX3C motif encoded by hGGT2 mRNAs disrupted autocleavage of the hGGT1 propeptide into a heterodimer, resulting in loss of plasma membrane localization and catalytic activity.
Innovation and conclusions: This is the first study to evaluate hGGT2 protein. The data show that hGGT2 does not encode a functional enzyme. Microarray data which have reported induction of hGGT2 mRNA should not be interpreted as induction of a protein that has a role in the metabolism of extracellular glutathione and in maintaining the redox status of the cell.
Figures


Similar articles
-
Novel insights into eukaryotic γ-glutamyltranspeptidase 1 from the crystal structure of the glutamate-bound human enzyme.J Biol Chem. 2013 Nov 1;288(44):31902-13. doi: 10.1074/jbc.M113.498139. Epub 2013 Sep 18. J Biol Chem. 2013. PMID: 24047895 Free PMC article.
-
Detection of distinct glycosylation patterns on human γ-glutamyl transpeptidase 1 using antibody-lectin sandwich array (ALSA) technology.BMC Biotechnol. 2014 Dec 6;14:101. doi: 10.1186/s12896-014-0101-0. BMC Biotechnol. 2014. PMID: 25479762 Free PMC article.
-
Gamma-glutamyltranspeptidase: disulfide bridges, propeptide cleavage, and activation in the endoplasmic reticulum.Methods Enzymol. 2005;401:426-49. doi: 10.1016/S0076-6879(05)01026-8. Methods Enzymol. 2005. PMID: 16399401 Review.
-
Human γ-Glutamyl Transpeptidase 1: STRUCTURES OF THE FREE ENZYME, INHIBITOR-BOUND TETRAHEDRAL TRANSITION STATES, AND GLUTAMATE-BOUND ENZYME REVEAL NOVEL MOVEMENT WITHIN THE ACTIVE SITE DURING CATALYSIS.J Biol Chem. 2015 Jul 10;290(28):17576-86. doi: 10.1074/jbc.M115.659680. Epub 2015 May 26. J Biol Chem. 2015. PMID: 26013825 Free PMC article.
-
Gene expression of gamma-glutamyltranspeptidase.Methods Enzymol. 2005;401:408-25. doi: 10.1016/S0076-6879(05)01025-6. Methods Enzymol. 2005. PMID: 16399400 Review.
Cited by
-
Metastable HIV-1 Surface Protein Env Sensitizes Cell Membranes to Transformation and Poration by Dual-Acting Virucidal Entry Inhibitors.Biochemistry. 2020 Feb 18;59(6):818-828. doi: 10.1021/acs.biochem.9b01008. Epub 2020 Jan 28. Biochemistry. 2020. PMID: 31942789 Free PMC article.
-
Novel insights into eukaryotic γ-glutamyltranspeptidase 1 from the crystal structure of the glutamate-bound human enzyme.J Biol Chem. 2013 Nov 1;288(44):31902-13. doi: 10.1074/jbc.M113.498139. Epub 2013 Sep 18. J Biol Chem. 2013. PMID: 24047895 Free PMC article.
-
Glutathione‑degrading enzymes in the complex landscape of tumors (Review).Int J Oncol. 2024 Jul;65(1):72. doi: 10.3892/ijo.2024.5660. Epub 2024 Jun 7. Int J Oncol. 2024. PMID: 38847236 Free PMC article. Review.
-
Sequence-Only Prediction of Super-Enhancers in Human Cell Lines Using Transformer Models.Biology (Basel). 2025 Feb 7;14(2):172. doi: 10.3390/biology14020172. Biology (Basel). 2025. PMID: 40001940 Free PMC article.
-
Non enzymatic upregulation of tissue factor expression by gamma-glutamyl transferase in human peripheral blood mononuclear cells.Thromb J. 2016 Nov 4;14:45. doi: 10.1186/s12959-016-0119-8. eCollection 2016. Thromb J. 2016. PMID: 27822142 Free PMC article.
References
-
- Baginski L. Tachon G. Falson F. Patton JS. Bakowsky U. Ehrhardt C. Reverse transcription polymerase chain reaction (RT-PCR) analysis of proteolytic enzymes in cultures of human respiratory epithelial cells. J Aerosol Med Pulm Drug Deliv. 2011;24:89–101. - PubMed
-
- Boanca G. Sand A. Barycki JJ. Uncoupling the enzymatic and autoprocessing activities of Helicobacter pylori gamma-glutamyltranspeptidase. J Biol Chem. 2006;281:19029–19037. - PubMed
-
- Brannigan JA. Dodson G. Duggleby HJ. Moody PC. Smith JL. Tomchick DR. Murzin AG. A protein catalytic framework with an N-terminal nucleophile is capable of self-activation. Nature. 1995;378:416–419. - PubMed
-
- Capraro MA. Hughey RP. Use of acivicin in the determination of rate constants for turnover of rat renal gamma-glutamyltranspeptidase. J Biol Chem. 1985;260:3408–3412. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

