Integrative genomic analysis of CREB defines a critical role for transcription factor networks in mediating the fed/fasted switch in liver
- PMID: 23682854
- PMCID: PMC3671974
- DOI: 10.1186/1471-2164-14-337
Integrative genomic analysis of CREB defines a critical role for transcription factor networks in mediating the fed/fasted switch in liver
Abstract
Background: Metabolic homeostasis in mammals critically depends on the regulation of fasting-induced genes by CREB in the liver. Previous genome-wide analysis has shown that only a small percentage of CREB target genes are induced in response to fasting-associated signaling pathways. The precise molecular mechanisms by which CREB specifically targets these genes in response to alternating hormonal cues remain to be elucidated.
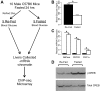
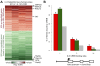
Results: We performed chromatin immunoprecipitation coupled to high-throughput sequencing of CREB in livers from both fasted and re-fed mice. In order to quantitatively compare the extent of CREB-DNA interactions genome-wide between these two physiological conditions we developed a novel, robust analysis method, termed the 'single sample independence' (SSI) test that greatly reduced the number of false-positive peaks. We found that CREB remains constitutively bound to its target genes in the liver regardless of the metabolic state. Integration of the CREB cistrome with expression microarrays of fasted and re-fed mouse livers and ChIP-seq data for additional transcription factors revealed that the gene expression switches between the two metabolic states are associated with co-localization of additional transcription factors at CREB sites.
Conclusions: Our results support a model in which CREB is constitutively bound to thousands of target genes, and combinatorial interactions between DNA-binding factors are necessary to achieve the specific transcriptional response of the liver to fasting. Furthermore, our genome-wide analysis identifies thousands of novel CREB target genes in liver, and suggests a previously unknown role for CREB in regulating ER stress genes in response to nutrient influx.
Figures



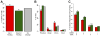

Similar articles
-
Transcriptional regulation of autophagy by an FXR-CREB axis.Nature. 2014 Dec 4;516(7529):108-11. doi: 10.1038/nature13949. Epub 2014 Nov 12. Nature. 2014. PMID: 25383523 Free PMC article.
-
Defining the CREB regulon: a genome-wide analysis of transcription factor regulatory regions.Cell. 2004 Dec 29;119(7):1041-54. doi: 10.1016/j.cell.2004.10.032. Cell. 2004. PMID: 15620361
-
Genome-wide gene expression and promoter binding analysis identifies NFIL3 as a repressor of C/EBP target genes in neuronal outgrowth.Mol Cell Neurosci. 2011 Feb;46(2):460-8. doi: 10.1016/j.mcn.2010.11.011. Epub 2010 Nov 26. Mol Cell Neurosci. 2011. PMID: 21112399
-
Coupling cAMP signaling to transcription in the liver: pivotal role of CREB and CREM.Exp Cell Res. 2002 May 1;275(2):143-54. doi: 10.1006/excr.2002.5491. Exp Cell Res. 2002. PMID: 11969286 Review.
-
Transcriptional and Chromatin Regulation during Fasting - The Genomic Era.Trends Endocrinol Metab. 2015 Dec;26(12):699-710. doi: 10.1016/j.tem.2015.09.005. Epub 2015 Oct 29. Trends Endocrinol Metab. 2015. PMID: 26520657 Free PMC article. Review.
Cited by
-
PCB126 Inhibits the Activation of AMPK-CREB Signal Transduction Required for Energy Sensing in Liver.Toxicol Sci. 2018 Jun 1;163(2):440-453. doi: 10.1093/toxsci/kfy041. Toxicol Sci. 2018. PMID: 29474705 Free PMC article.
-
CrossHub: a tool for multi-way analysis of The Cancer Genome Atlas (TCGA) in the context of gene expression regulation mechanisms.Nucleic Acids Res. 2016 Apr 20;44(7):e62. doi: 10.1093/nar/gkv1478. Epub 2016 Jan 14. Nucleic Acids Res. 2016. PMID: 26773058 Free PMC article.
-
Transcriptional regulatory logic of the diurnal cycle in the mouse liver.PLoS Biol. 2017 Apr 17;15(4):e2001069. doi: 10.1371/journal.pbio.2001069. eCollection 2017 Apr. PLoS Biol. 2017. PMID: 28414715 Free PMC article.
-
Transcriptional Regulation of ZNF638 in Thermogenic Cells by the cAMP Response Element Binding Protein in Male Mice.J Endocr Soc. 2019 Oct 3;3(12):2326-2340. doi: 10.1210/js.2019-00238. eCollection 2019 Dec 1. J Endocr Soc. 2019. PMID: 31745529 Free PMC article.
-
Tissue-specific BMAL1 cistromes reveal that rhythmic transcription is associated with rhythmic enhancer-enhancer interactions.Genes Dev. 2019 Mar 1;33(5-6):294-309. doi: 10.1101/gad.322198.118. Epub 2019 Feb 25. Genes Dev. 2019. PMID: 30804225 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

