Quantitative phosphoproteomics reveals extensive cellular reprogramming during HIV-1 entry
- PMID: 23684312
- PMCID: PMC4104530
- DOI: 10.1016/j.chom.2013.04.011
Quantitative phosphoproteomics reveals extensive cellular reprogramming during HIV-1 entry
Abstract
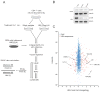
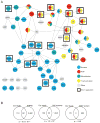
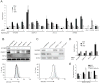
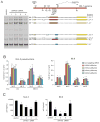
Receptor engagement by HIV-1 during host cell entry activates signaling pathways that can reprogram the cell for optimal viral replication. To obtain a global view of the signaling events induced during HIV-1 entry, we conducted a quantitative phosphoproteomics screen of primary human CD4(+) T cells after infection with an HIV-1 strain that engages the receptors CD4 and CXCR4. We quantified 1,757 phosphorylation sites with high stringency. The abundance of 239 phosphorylation sites from 175 genes, including several proteins in pathways known to be impacted by HIV-receptor binding, changed significantly within a minute after HIV-1 exposure. Several previously uncharacterized HIV-1 host factors were also identified and confirmed through RNAi depletion studies. Surprisingly, five serine/arginine-rich (SR) proteins involved in messenger RNA splicing, including the splicing factor SRm300 (SRRM2), were differentially phosophorylated. Mechanistic studies with SRRM2 suggest that HIV-1 modulates host cell alternative splicing machinery during entry in order to facilitate virus replication and release.
Copyright © 2013 Elsevier Inc. All rights reserved.
Figures

Similar articles
-
Global phosphoproteomics of CCR5-tropic HIV-1 signaling reveals reprogramming of cellular protein production pathways and identifies p70-S6K1 and MK2 as HIV-responsive kinases required for optimal infection of CD4+ T cells.Retrovirology. 2018 Jul 3;15(1):44. doi: 10.1186/s12977-018-0423-4. Retrovirology. 2018. PMID: 29970186 Free PMC article.
-
The Role of Integrin α4β7 Signaling in Human Immunodeficiency Virus-1 Pathogenesis and Viral Entry in Primary CD4+ T Cells As Revealed by Comparative Phosphoproteomic Signatures.OMICS. 2020 Jul;24(7):437-450. doi: 10.1089/omi.2019.0196. Epub 2020 Jun 9. OMICS. 2020. PMID: 32522079
-
Quantitative Temporal Viromics of an Inducible HIV-1 Model Yields Insight to Global Host Targets and Phospho-Dynamics Associated with Protein Vpr.Mol Cell Proteomics. 2017 Aug;16(8):1447-1461. doi: 10.1074/mcp.M116.066019. Epub 2017 Jun 12. Mol Cell Proteomics. 2017. PMID: 28606917 Free PMC article.
-
Human Immunodeficiency Virus Type 1 Cellular Entry and Exit in the T Lymphocytic and Monocytic Compartments: Mechanisms and Target Opportunities During Viral Disease.Adv Virus Res. 2015;93:257-311. doi: 10.1016/bs.aivir.2015.04.001. Epub 2015 May 8. Adv Virus Res. 2015. PMID: 26111588 Review.
-
The molecular basis of HIV entry.Cell Microbiol. 2012 Aug;14(8):1183-92. doi: 10.1111/j.1462-5822.2012.01812.x. Epub 2012 Jun 5. Cell Microbiol. 2012. PMID: 22583677 Free PMC article. Review.
Cited by
-
An atlas of posttranslational modifications on RNA binding proteins.Nucleic Acids Res. 2022 May 6;50(8):4329-4339. doi: 10.1093/nar/gkac243. Nucleic Acids Res. 2022. PMID: 35438783 Free PMC article.
-
Bioinformatics and Connectivity Map Analysis Suggest Viral Infection as a Critical Causative Factor of Hashimoto's Thyroiditis.Int J Mol Sci. 2023 Jan 6;24(2):1157. doi: 10.3390/ijms24021157. Int J Mol Sci. 2023. PMID: 36674671 Free PMC article.
-
HIV and innate immunity - a genomics perspective.F1000Prime Rep. 2013 Aug 1;5:29. doi: 10.12703/P5-29. eCollection 2013. F1000Prime Rep. 2013. PMID: 23967380 Free PMC article.
-
HIV signaling through CD4 and CCR5 activates Rho family GTPases that are required for optimal infection of primary CD4+ T cells.Retrovirology. 2017 Jan 24;14(1):4. doi: 10.1186/s12977-017-0328-7. Retrovirology. 2017. PMID: 28114951 Free PMC article.
-
Improving data quality and preserving HCD-generated reporter ions with EThcD for isobaric tag-based quantitative proteomics and proteome-wide PTM studies.Anal Chim Acta. 2017 May 22;968:40-49. doi: 10.1016/j.aca.2017.03.003. Epub 2017 Mar 16. Anal Chim Acta. 2017. PMID: 28395773 Free PMC article.
References
-
- Balabanian K, Harriague J, Decrion C, Lagane B, Shorte S, Baleux F, Virelizier JL, Arenzana-Seisdedos F, Chakrabarti LA. CXCR4-tropic HIV-1 envelope glycoprotein functions as a viral chemokine in unstimulated primary CD4+ T lymphocytes. J Immunol. 2004;173:7150–7160. - PubMed
-
- Barrero-Villar M, Cabrero JR, Gordon-Alonso M, Barroso-Gonzalez J, Alvarez-Losada S, Munoz-Fernandez MA, Sanchez-Madrid F, Valenzuela-Fernandez A. Moesin is required for HIV-1-induced CD4-CXCR4 interaction, F-actin redistribution, membrane fusion and viral infection in lymphocytes. J Cell Sci. 2009;122:103–113. - PubMed
-
- Brass AL, Dykxhoorn DM, Benita Y, Yan N, Engelman A, Xavier RJ, Lieberman J, Elledge SJ. Identification of host proteins required for HIV infection through a functional genomic screen. Science. 2008;319:921–926. - PubMed
-
- Briant L, Coudronniere N, Robert-Hebmann V, Benkirane M, Devaux C. Binding of HIV-1 virions or gp120-anti-gp120 immune complexes to HIV-1-infected quiescent peripheral blood mononuclear cells reveals latent infection. J Immunol. 1996;156:3994–4004. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

