Transcriptional repression of Gata3 is essential for early B cell commitment
- PMID: 23684985
- PMCID: PMC3664383
- DOI: 10.1016/j.immuni.2013.01.014
Transcriptional repression of Gata3 is essential for early B cell commitment
Abstract
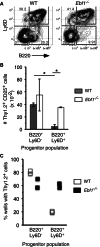
The mechanisms underlying the silencing of alternative fate potentials in very early B cell precursors remain unclear. Using gain- and loss-of-function approaches together with a synthetic Zinc-finger polypeptide (6ZFP) engineered to prevent transcription factor binding to a defined cis element, we show that the transcription factor EBF1 promotes B cell lineage commitment by directly repressing expression of the T-cell-lineage-requisite Gata3 gene. Ebf1-deficient lymphoid progenitors exhibited increased T cell lineage potential and elevated Gata3 transcript expression, whereas enforced EBF1 expression inhibited T cell differentiation and caused rapid loss of Gata3 mRNA. Notably, 6ZFP-mediated perturbation of EBF1 binding to a Gata3 regulatory region restored Gata3 expression, abrogated EBF1-driven suppression of T cell differentiation, and prevented B cell differentiation via a GATA3-dependent mechanism. Furthermore, EBF1 binding to Gata3 regulatory sites induced repressive histone modifications across this region. These data identify a transcriptional circuit critical for B cell lineage commitment.
Copyright © 2013 Elsevier Inc. All rights reserved.
Figures





References
-
- Allman D., Sambandam A., Kim S., Miller J.P., Pagan A., Well D., Meraz A., Bhandoola A. Thymopoiesis independent of common lymphoid progenitors. Nat. Immunol. 2003;4:168–174. - PubMed
-
- Barski A., Cuddapah S., Cui K., Roh T.Y., Schones D.E., Wang Z., Wei G., Chepelev I., Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

