Identification of functional modules of AKMT, a novel lysine methyltransferase regulating the motility of Toxoplasma gondii
- PMID: 23685344
- PMCID: PMC3740173
- DOI: 10.1016/j.molbiopara.2013.05.004
Identification of functional modules of AKMT, a novel lysine methyltransferase regulating the motility of Toxoplasma gondii
Abstract
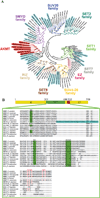
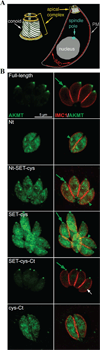
The intracellular parasite Toxoplasma gondii is a leading cause of congenital neurological defects. To cause disease, it must reiterate its lytic cycle through host cell invasion, replication, and parasite egress. This requires the parasite to sense changes in its environment and switch between the non-motile (for replication) and motile (for invasion and egress) states appropriately. Recently, we discovered a previously unknown mechanism of motility regulation in T. gondii, mediated by a lysine methyltransferase, AKMT (for Apical complex lysine (K) methyltransferase). When AKMT is absent, activation of motility is inhibited, which compromises parasite invasion and egress, and thus severely impairs the lytic cycle. Although the methyltransferase activity of AKMT has been established, the phylogenetic relationship of AKMT with other better studied lysine methyltransferases (KMTs) was not known. Also unknown was the functional relationships between different domains of AKMT. In this work we carried out phylogenetic analyses, which show that AKMT orthologs form a new subfamily of KMTs. We systematically generated truncation mutants of AKMT, and discovered that the predicted enzymatic domain alone is a very poor enzyme and cannot complement the function of AKMT in vivo. Interestingly, the N- and C-terminal domains of the AKMT have drastically different impacts on its enzyme activity, localization as well as in vivo function. Our results thus reveal that AKMT is an unusual, parasite-specific enzyme and identified regions and interactions within this novel lysine methyltransferase that can be used as drug targets.
Copyright © 2013 Elsevier B.V. All rights reserved.
Figures




References
-
- Levine ND. Progress in taxonomy of the Apicomplexan protozoa. J Protozool. 1988;35:518–520. - PubMed
-
- Holliman RE. Toxoplasmosis and the acquired immune deficiency syndrome. J Infect. 1988;16:121–128. - PubMed
-
- Fleming AF. Opportunistic infections in AIDS in developed and developing countries. Trans R Soc Trop Med Hyg. 1990;84(Suppl 1):1–6. - PubMed
-
- Belanger F, Derouin F, Grangeot-Keros L, Meyer L. Incidence and risk factors of toxoplasmosis in a cohort of human immunodeficiency virus-infected patients: 1988–1995. HEMOCO and SEROCO Study Groups. Clin Infect Dis. 1999;28:575–581. - PubMed
-
- Vastagh I, Jelencsik I, Barsi P, Balint K, Szlavik J, Szirmai I. Single cerebral Toxoplasma abscess: first manifestation of HIV infection. Eur J Neurol. 1999;6:725–726. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

