Recent thymic emigrants and mature naive T cells exhibit differential DNA methylation at key cytokine loci
- PMID: 23686491
- PMCID: PMC3679312
- DOI: 10.4049/jimmunol.1300181
Recent thymic emigrants and mature naive T cells exhibit differential DNA methylation at key cytokine loci
Abstract
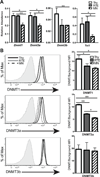
Recent thymic emigrants (RTEs) are the youngest T cells in the lymphoid periphery and exhibit phenotypic and functional characteristics distinct from those of their more mature counterparts in the naive peripheral T cell pool. We show in this study that the Il2 and Il4 promoter regions of naive CD4(+) RTEs are characterized by site-specific hypermethylation compared with those of both mature naive (MN) T cells and the thymocyte precursors of RTEs. Thus, RTEs do not merely occupy a midpoint between the thymus and the mature T cell pool, but represent a distinct transitional T cell population. Furthermore, RTEs and MN T cells exhibit distinct CpG DNA methylation patterns both before and after activation. Compared with MN T cells, RTEs express higher levels of several enzymes that modify DNA methylation, and inhibiting methylation during culture allows RTEs to reach MN T cell levels of cytokine production. Collectively, these data suggest that the functional differences that distinguish RTEs from MN T cells are influenced by epigenetic mechanisms and provide clues to a mechanistic basis for postthymic maturation.
Figures



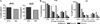

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

