Antisense oligonucleotides: treating neurodegeneration at the level of RNA
- PMID: 23686823
- PMCID: PMC3701770
- DOI: 10.1007/s13311-013-0194-5
Antisense oligonucleotides: treating neurodegeneration at the level of RNA
Abstract
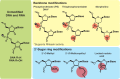
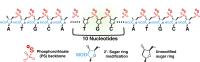
Adequate therapies are lacking for Alzheimer's disease, Parkinson's disease, Huntington's disease, amyotrophic lateral sclerosis, and other neurodegenerative diseases. The ability to use antisense oligonucleotides (ASOs) to target disease-associated genes by means of RNA may offer a potent approach for the treatment of these, and other, neurodegenerative disorders. In modifying the basic backbone chemistry, chemical groups, and target sequence, ASOs can act through numerous mechanisms to decrease or increase total protein levels, preferentially shift splicing patterns, and inhibit microRNAs, all at the level of the RNA molecule. Here, we discuss many of the more commonly used ASO chemistries, as well as the different mechanisms of action that can result from these specific chemical modifications. When applied to multiple neurodegenerative mouse models, ASOs that specifically target the detrimental transgenes have been shown to rescue disease associated phenotypes in vivo. These supporting mouse model data have moved the ASOs from the bench to the clinic, with two neuro-focused human clinical trials now underway and several more being proposed. Although still early in development, translating ASOs into human patients for neurodegeneration appears promising.
Figures


References
-
- Eckstein F. Phosphorothioate oligodeoxynucleotides: what is their origin and what is unique about them? Antisense Nucleic Acid Drug Dev. 2000;10:117–121. - PubMed
-
- Brown D, Kang S, Gryaznov S, DeDionisio L, Heidenreich O, Sullivan S, et al. Effect of phosphorothioate modification of oligodeoxynucleotides on specific protein binding. J Biol Chem. 1994;269:26801–26805. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

