Toward creation of a cancer drug toxicity knowledge base: automatically extracting cancer drug-side effect relationships from the literature
- PMID: 23686935
- PMCID: PMC3912715
- DOI: 10.1136/amiajnl-2012-001584
Toward creation of a cancer drug toxicity knowledge base: automatically extracting cancer drug-side effect relationships from the literature
Abstract
Objective: A comprehensive and machine-understandable cancer drug-side effect (drug-SE) relationship knowledge base is important for in silico cancer drug target discovery, drug repurposing, and toxicity predication, and for personalized risk-benefit decisions by cancer patients. While US Food and Drug Administration (FDA) drug labels capture well-known cancer drug SE information, much cancer drug SE knowledge remains buried the published biomedical literature. We present a relationship extraction approach to extract cancer drug-SE pairs from the literature.
Data and methods: We used 21,354,075 MEDLINE records as the text corpus. We extracted drug-SE co-occurrence pairs using a cancer drug lexicon and a clean SE lexicon that we created. We then developed two filtering approaches to remove drug-disease treatment pairs and subsequently a ranking scheme to further prioritize filtered pairs. Finally, we analyzed relationships among SEs, gene targets, and indications.
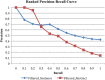
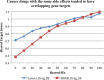
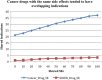
Results: We extracted 56,602 cancer drug-SE pairs. The filtering algorithms improved the precision of extracted pairs from 0.252 at baseline to 0.426, representing a 69% improvement in precision with no decrease in recall. The ranking algorithm further prioritized filtered pairs and achieved a precision of 0.778 for top-ranked pairs. We showed that cancer drugs that share SEs tend to have overlapping gene targets and overlapping indications.
Conclusions: The relationship extraction approach is effective in extracting many cancer drug-SE pairs from the literature. This unique knowledge base, when combined with existing cancer drug SE knowledge, can facilitate drug target discovery, drug repurposing, and toxicity prediction.
Keywords: Cancer Drug Toxicity; Drug Repurposing; Drug Target Discovery; Information Extraction; Natural Language Processing; Text Mining.
Figures


References
-
- Ladewski LA, Belknap SM, Nebeker JR, et al. Dissemination of information on potentially fatal adverse drug reactions for cancer drugs from 2000 to 2002: first results from the research on adverse drug events and reports project. J Clin Oncol 2003;21:3859–66 - PubMed
-
- Campillos M, Kuhn M, Gavin AC, et al. Drug target identification using side-effect similarity. Science 2008;321:263–6 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

