Targeted glycoprotein enrichment and identification in stromal cell secretomes using azido sugar metabolic labeling
- PMID: 23687070
- PMCID: PMC4331031
- DOI: 10.1002/prca.201300006
Targeted glycoprotein enrichment and identification in stromal cell secretomes using azido sugar metabolic labeling
Abstract
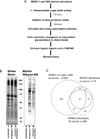
Purpose: Effectively identifying the proteins present in the cellular secretome is complicated due to the presence of cellular protein leakage and serum protein supplements in culture media. A metabolic labeling and click chemistry capture method is described that facilitates the detection of lower abundance glycoproteins in the secretome, even in the presence of serum.
Experimental design: Two stromal cell lines were incubated with tetraacetylated sugar-azide analogs for 48 h in serum-free and low-serum conditions. Sugar-azide labeled glycoproteins were covalently linked to alkyne-beads, followed by on-bead trypsin digestion and MS/MS. The resulting glycoproteins were compared between media conditions, cell lines, and azide-sugar labels.
Results: Alkyne-bead capture of sugar-azide modified glycoproteins in stromal cell culture media significantly improved the detection of lower abundance secreted glycoproteins compared to standard serum-free secretome preparations. Over 100 secreted glycoproteins were detected in each stromal cell line and significantly enriched relative to a standard secretome preparation.
Conclusion and clinical relevance: Sugar-azide metabolic labeling is an effective way to enrich for secreted glycoproteins present in cell line secretomes, even in culture media supplemented with serum. The method has utility for identifying secreted stromal proteins associated with cancer progression and the epithelial-to-mesenchymal transition.
© 2013 WILEY-VCH Verlag GmbH & Co. KGaA, Weinheim.
Conflict of interest statement
The authors have declared no conflict of interest.
Figures

Similar articles
-
[Comparison of the performance of secretome analysis based on metabolic labeling by three unnatural sugars].Se Pu. 2021 Oct;39(10):1086-1093. doi: 10.3724/SP.J.1123.2021.04017. Se Pu. 2021. PMID: 34505430 Free PMC article. Chinese.
-
Metabolic labeling and click chemistry detection of glycoprotein markers of mesenchymal stem cell differentiation.Methods Mol Biol. 2011;698:459-84. doi: 10.1007/978-1-60761-999-4_33. Methods Mol Biol. 2011. PMID: 21431538
-
From mechanism to mouse: a tale of two bioorthogonal reactions.Acc Chem Res. 2011 Sep 20;44(9):666-76. doi: 10.1021/ar200148z. Epub 2011 Aug 15. Acc Chem Res. 2011. PMID: 21838330 Free PMC article.
-
Glycocapture-based proteomics for secretome analysis.Proteomics. 2013 Feb;13(3-4):512-25. doi: 10.1002/pmic.201200414. Epub 2013 Jan 14. Proteomics. 2013. PMID: 23197367 Review.
-
Synthesis of Porphyrin, Chlorin and Phthalocyanine Derivatives by Azide-Alkyne Click Chemistry.Curr Med Chem. 2015;22(28):3217-54. doi: 10.2174/0929867322666150716115832. Curr Med Chem. 2015. PMID: 26179994 Review.
Cited by
-
Intact glycopeptide characterization using mass spectrometry.Expert Rev Proteomics. 2016 May;13(5):513-22. doi: 10.1586/14789450.2016.1172965. Expert Rev Proteomics. 2016. PMID: 27140194 Free PMC article. Review.
-
MALDI imaging mass spectrometry profiling of proteins and lipids in clear cell renal cell carcinoma.Proteomics. 2014 Apr;14(7-8):924-35. doi: 10.1002/pmic.201300434. Epub 2014 Mar 3. Proteomics. 2014. PMID: 24497498 Free PMC article.
-
Enhancing Comprehensive Analysis of Secreted Glycoproteins from Cultured Cells without Serum Starvation.Anal Chem. 2021 Feb 2;93(4):2694-2705. doi: 10.1021/acs.analchem.0c05126. Epub 2021 Jan 4. Anal Chem. 2021. PMID: 33397101 Free PMC article.
-
Advances in mass spectrometry-based glycoproteomics.Electrophoresis. 2018 Dec;39(24):3104-3122. doi: 10.1002/elps.201800272. Epub 2018 Oct 9. Electrophoresis. 2018. PMID: 30203847 Free PMC article. Review.
-
Chemical Glycoproteomics.Chem Rev. 2016 Dec 14;116(23):14277-14306. doi: 10.1021/acs.chemrev.6b00023. Epub 2016 Nov 18. Chem Rev. 2016. PMID: 27960262 Free PMC article. Review.
References
-
- Makridakis M, Vlahou A. Secretome proteomics for discovery of cancer biomarkers. J. Proteomics. 2010;73:2291–2305. - PubMed
-
- Kulasingam V, Diamandis EP. Strategies for discoveringnovel cancer biomarkers through utilization of emerging technologies. Nat. Clin. Pract. Oncol. 2008;5:588–699. - PubMed
-
- May M. From cells, secrets of the secretome leak out. Nat. Med. 2009;15:828. - PubMed
-
- Drake RR, Schwegler EE, Malik G, Diaz J, et al. Lectin capture strategies combined withmass spectrometry for the discovery of serum glycoprotein biomarkers. Mol. Cell. Proteomics. 2006;5:1957–1967. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

