PCR on yeast colonies: an improved method for glyco-engineered Saccharomyces cerevisiae
- PMID: 23688076
- PMCID: PMC3664073
- DOI: 10.1186/1756-0500-6-201
PCR on yeast colonies: an improved method for glyco-engineered Saccharomyces cerevisiae
Abstract
Background: Saccharomyces cerevisiae is extensively used in bio-industries. However, its genetic engineering to introduce new metabolism pathways can cause unexpected phenotypic alterations. For example, humanisation of the glycosylation pathways is a high priority pharmaceutical industry goal for production of therapeutic glycoproteins in yeast. Genomic modifications can lead to several described physiological changes: biomass yields decrease, temperature sensitivity or cell wall structure modifications. We have observed that deletion of several N-mannosyltransferases in Saccharomyces cerevisiae, results in strains that can no longer be analyzed by classical PCR on yeast colonies.
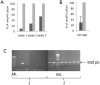
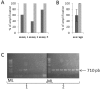
Findings: In order to validate our glyco-engineered Saccharomyces cerevisiae strains, we developed a new protocol to carry out PCR directly on genetically modified yeast colonies. A liquid culture phase, combined with the use of a Hot Start DNA polymerase, allows a 3-fold improvement of PCR efficiency. The results obtained are repeatable and independent of the targeted sequence; as such the protocol is well adapted for intensive screening applications.
Conclusions: The developed protocol enables by-passing of many of the difficulties associated with PCR caused by phenotypic modifications brought about by humanisation of the glycosylation in yeast and allows rapid validation of glyco-engineered Saccharomyces cerevisiae cells. It has the potential to be extended to other yeast strains presenting cell wall structure modifications.
Figures


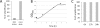
Similar articles
-
Essential role of YlMPO1, a novel Yarrowia lipolytica homologue of Saccharomyces cerevisiae MNN4, in mannosylphosphorylation of N- and O-linked glycans.Appl Environ Microbiol. 2011 Feb;77(4):1187-95. doi: 10.1128/AEM.02323-10. Epub 2010 Dec 23. Appl Environ Microbiol. 2011. PMID: 21183647 Free PMC article.
-
[Reconstruction of N-glycosylation pathway for producing human glycoproteins in Saccharomyces cerevisiae].Wei Sheng Wu Xue Bao. 2014 May 4;54(5):509-16. Wei Sheng Wu Xue Bao. 2014. PMID: 25199249 Chinese.
-
Engineering yeast for producing human glycoproteins: where are we now?Future Microbiol. 2015;10(1):21-34. doi: 10.2217/fmb.14.104. Future Microbiol. 2015. PMID: 25598335 Free PMC article.
-
The use of genetically modified Saccharomyces cerevisiae strains in the wine industry.Appl Microbiol Biotechnol. 2005 Aug;68(3):292-304. doi: 10.1007/s00253-005-1994-2. Epub 2005 Apr 26. Appl Microbiol Biotechnol. 2005. PMID: 15856224 Review.
-
[Development and application of Saccharomyces cerevisiae cell-surface display for bioethanol production].Sheng Wu Gong Cheng Xue Bao. 2012 Aug;28(8):901-11. Sheng Wu Gong Cheng Xue Bao. 2012. PMID: 23185890 Review. Chinese.
Cited by
-
Bacteria and Yeast Colony PCR.Methods Mol Biol. 2023;2967:209-221. doi: 10.1007/978-1-0716-3358-8_17. Methods Mol Biol. 2023. PMID: 37608114
-
Heterologous Expression of Recombinant Ginseng Tetradecapeptide in Saccharomyces cerevisiae and Evaluation of Its Biological Activity.Foods. 2025 Jun 10;14(12):2049. doi: 10.3390/foods14122049. Foods. 2025. PMID: 40565658 Free PMC article.
-
Rapid and robust squashed spore/colony PCR of industrially important fungi.Fungal Biol Biotechnol. 2023 Jul 8;10(1):15. doi: 10.1186/s40694-023-00163-0. Fungal Biol Biotechnol. 2023. PMID: 37422681 Free PMC article.
-
Yeast Expression Systems: Overview and Recent Advances.Mol Biotechnol. 2019 May;61(5):365-384. doi: 10.1007/s12033-019-00164-8. Mol Biotechnol. 2019. PMID: 30805909 Review.
-
Bootleg Biology: a Semester-Long CURE Using Wild Yeast to Brew Beer.J Microbiol Biol Educ. 2022 Oct 6;23(3):e00336-21. doi: 10.1128/jmbe.00336-21. eCollection 2022 Dec. J Microbiol Biol Educ. 2022. PMID: 36532218 Free PMC article.
References
-
- Microorganisms & Microbial-Derived Ingredients Used in Food. http://www.fda.gov/Food/FoodIngredientsPackaging/ucm078956.htm.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

