PCR on yeast colonies: an improved method for glyco-engineered Saccharomyces cerevisiae
- PMID: 23688076
- PMCID: PMC3664073
- DOI: 10.1186/1756-0500-6-201
PCR on yeast colonies: an improved method for glyco-engineered Saccharomyces cerevisiae
Abstract
Background: Saccharomyces cerevisiae is extensively used in bio-industries. However, its genetic engineering to introduce new metabolism pathways can cause unexpected phenotypic alterations. For example, humanisation of the glycosylation pathways is a high priority pharmaceutical industry goal for production of therapeutic glycoproteins in yeast. Genomic modifications can lead to several described physiological changes: biomass yields decrease, temperature sensitivity or cell wall structure modifications. We have observed that deletion of several N-mannosyltransferases in Saccharomyces cerevisiae, results in strains that can no longer be analyzed by classical PCR on yeast colonies.
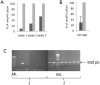
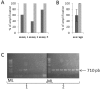
Findings: In order to validate our glyco-engineered Saccharomyces cerevisiae strains, we developed a new protocol to carry out PCR directly on genetically modified yeast colonies. A liquid culture phase, combined with the use of a Hot Start DNA polymerase, allows a 3-fold improvement of PCR efficiency. The results obtained are repeatable and independent of the targeted sequence; as such the protocol is well adapted for intensive screening applications.
Conclusions: The developed protocol enables by-passing of many of the difficulties associated with PCR caused by phenotypic modifications brought about by humanisation of the glycosylation in yeast and allows rapid validation of glyco-engineered Saccharomyces cerevisiae cells. It has the potential to be extended to other yeast strains presenting cell wall structure modifications.
Figures


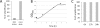
References
-
- Microorganisms & Microbial-Derived Ingredients Used in Food. http://www.fda.gov/Food/FoodIngredientsPackaging/ucm078956.htm.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

