Genomics-driven discovery of the pneumocandin biosynthetic gene cluster in the fungus Glarea lozoyensis
- PMID: 23688303
- PMCID: PMC3672099
- DOI: 10.1186/1471-2164-14-339
Genomics-driven discovery of the pneumocandin biosynthetic gene cluster in the fungus Glarea lozoyensis
Abstract
Background: The antifungal therapy caspofungin is a semi-synthetic derivative of pneumocandin B0, a lipohexapeptide produced by the fungus Glarea lozoyensis, and was the first member of the echinocandin class approved for human therapy. The nonribosomal peptide synthetase (NRPS)-polyketide synthases (PKS) gene cluster responsible for pneumocandin biosynthesis from G. lozoyensis has not been elucidated to date. In this study, we report the elucidation of the pneumocandin biosynthetic gene cluster by whole genome sequencing of the G. lozoyensis wild-type strain ATCC 20868.
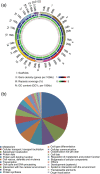
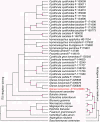
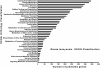
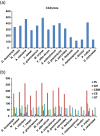
Results: The pneumocandin biosynthetic gene cluster contains a NRPS (GLNRPS4) and a PKS (GLPKS4) arranged in tandem, two cytochrome P450 monooxygenases, seven other modifying enzymes, and genes for L-homotyrosine biosynthesis, a component of the peptide core. Thus, the pneumocandin biosynthetic gene cluster is significantly more autonomous and organized than that of the recently characterized echinocandin B gene cluster. Disruption mutants of GLNRPS4 and GLPKS4 no longer produced the pneumocandins (A0 and B0), and the Δglnrps4 and Δglpks4 mutants lost antifungal activity against the human pathogenic fungus Candida albicans. In addition to pneumocandins, the G. lozoyensis genome encodes a rich repertoire of natural product-encoding genes including 24 PKSs, six NRPSs, five PKS-NRPS hybrids, two dimethylallyl tryptophan synthases, and 14 terpene synthases.
Conclusions: Characterization of the gene cluster provides a blueprint for engineering new pneumocandin derivatives with improved pharmacological properties. Whole genome estimation of the secondary metabolite-encoding genes from G. lozoyensis provides yet another example of the huge potential for drug discovery from natural products from the fungal kingdom.
Figures





References
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

