Pseudomonas fluorescens SBW25 produces furanomycin, a non-proteinogenic amino acid with selective antimicrobial properties
- PMID: 23688329
- PMCID: PMC3662646
- DOI: 10.1186/1471-2180-13-111
Pseudomonas fluorescens SBW25 produces furanomycin, a non-proteinogenic amino acid with selective antimicrobial properties
Erratum in
- BMC Microbiol. 2013;13:263
Abstract
Background: Pseudomonas fluorescens SBW25 has been extensively studied because of its plant growth promoting properties and potential as a biocontrol agent. The genome of SBW25 has been sequenced, and among sequenced strains of pseudomonads, SBW25 appears to be most closely related to P. fluorescens WH6. In the authors' laboratories, WH6 was previously shown to produce and secrete 4-formylaminooxyvinylglycine (FVG), a non-proteinogenic amino acid with selective herbicidal and antimicrobial activity. Although SBW25 does not have the genetic capacity to produce FVG, we were interested in determining whether this pseudomonad might produce some other type of non-proteinogenic amino acid.
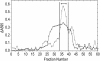
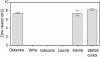
Results: P. fluorescens SBW25 was found to produce and secrete a ninhydrin-reactive compound with selective antimicrobial properties. This compound was purified from SBW25 culture filtrate and identified as the non-proteinogenic amino acid L-furanomycin [2S,2'R,5'S)-2-amino-2-(5'methyl-2',5'-dihydrofuran-2'-yl)acetic acid].
Conclusions: The identification of furanomycin as a secondary metabolite of SBW25 is the first report of the production of furanomycin by a pseudomonad. This compound was known previously only as a natural product produced by a strain of Streptomyces. This report adds furanomycin to the small list of non-proteinogenic amino acids that have been identified as secondary products of pseudomonads. This study also extends the list of bacteria that are inhibited by furanomycin to include several plant pathogenic bacteria.
Figures



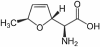
Similar articles
-
Genome-based discovery, structure prediction and functional analysis of cyclic lipopeptide antibiotics in Pseudomonas species.Mol Microbiol. 2007 Jan;63(2):417-28. doi: 10.1111/j.1365-2958.2006.05525.x. Mol Microbiol. 2007. PMID: 17241198
-
Genomic, genetic and structural analysis of pyoverdine-mediated iron acquisition in the plant growth-promoting bacterium Pseudomonas fluorescens SBW25.BMC Microbiol. 2008 Jan 14;8:7. doi: 10.1186/1471-2180-8-7. BMC Microbiol. 2008. PMID: 18194565 Free PMC article.
-
An improved, high-quality draft genome sequence of the Germination-Arrest Factor-producing Pseudomonas fluorescens WH6.BMC Genomics. 2010 Sep 28;11:522. doi: 10.1186/1471-2164-11-522. BMC Genomics. 2010. PMID: 20920191 Free PMC article.
-
Helical Antimicrobial Peptide Foldamers Containing Non-proteinogenic Amino Acids.ChemMedChem. 2021 Apr 20;16(8):1226-1233. doi: 10.1002/cmdc.202000940. Epub 2021 Feb 10. ChemMedChem. 2021. PMID: 33565721 Review.
-
Naturally occurring amino acid derivatives with herbicidal, fungicidal or insecticidal activity.Amino Acids. 2016 Apr;48(4):929-940. doi: 10.1007/s00726-016-2176-5. Epub 2016 Jan 22. Amino Acids. 2016. PMID: 26801938 Review.
Cited by
-
Interstrain interactions between bacteria isolated from vacuum-packaged refrigerated beef.Appl Environ Microbiol. 2015 Apr;81(8):2753-61. doi: 10.1128/AEM.03933-14. Epub 2015 Feb 6. Appl Environ Microbiol. 2015. PMID: 25662972 Free PMC article.
-
Unexpected distribution of the 4-formylaminooxyvinylglycine (FVG) biosynthetic pathway in Pseudomonas and beyond.PLoS One. 2021 Apr 23;16(4):e0247348. doi: 10.1371/journal.pone.0247348. eCollection 2021. PLoS One. 2021. PMID: 33891610 Free PMC article.
-
Potential of Pantoea dispersa as an effective biocontrol agent for black rot in sweet potato.Sci Rep. 2019 Nov 8;9(1):16354. doi: 10.1038/s41598-019-52804-3. Sci Rep. 2019. PMID: 31704990 Free PMC article.
-
Stable, fluorescent markers for tracking synthetic communities and assembly dynamics.Microbiome. 2024 May 7;12(1):81. doi: 10.1186/s40168-024-01792-2. Microbiome. 2024. PMID: 38715147 Free PMC article.
-
Microbial bioformulation: a microbial assisted biostimulating fertilization technique for sustainable agriculture.Front Plant Sci. 2023 Dec 12;14:1270039. doi: 10.3389/fpls.2023.1270039. eCollection 2023. Front Plant Sci. 2023. PMID: 38148858 Free PMC article. Review.
References
-
- Bossis E, Lemanceau P, Latour X, Gardan L. The taxonomy of Pseudomonas fluorescens and Pseudomonas putida: current status and need for revision. Agronomie. 2000;20:51–63.
-
- Yamamoto S, Kasai H, Arnold DL, Jackson RW, Vivian A, Harayama S. Phylogeny of the genus Pseudomonas: intrageneric structure reconstructed from the nucleotide sequences of gyrB and rpoD genes. Microbiol. 2000;146:2385–2394. - PubMed
-
- Silby MW, Cerdñeo-Tárraga AM, Vernikos GS, Giddens SR, Jackson RW, Preston GM, Zhang X-X, Moon CD, Gehrig SM, Godfrey SAC, Knight CG, Malone JG, Robinson Z, Spiers AJ, Harris S, Challis GL, Yaxley AM, Harris D, Seeger K, Murphy L, Rutter S, Squares R, Quail MA, Saunders E, Mavromatis K, Brettin TS, Bentley SD, Hothersall J, Stephens E, Thomas CM, Parkhill J, Levy SB, Rainey PB, Thomson NR. Genomic and genetic analyses of diversity and plant interactions of Pseudomonas fluorescens. Genome Biol. 2009;10:R51. doi: 10.1186/gb-2009-10-5-r51. - DOI - PMC - PubMed
-
- Loper JE, Hassan KA, Mavrodi DV, Davis EW II, Lim CK, Shaffer BT, Elbourne LD, Stockwell VO, Hartney SL, Breakwell K, Henkels MD, Tetu SG, Rangel LI, Kidarsa TA, Wilson NL, van de Mortel JE, Song C, Blumhagen R, Radune D, Hostetler JB, Brinkac LM, Durkin AS, Kluepfel DA, Wechter WP, Anderson AJ, Kim YC, Pierson LS III, Pierson EA, Lindow SE, Kobayashi DY, Raaijmakers JM, Weller DM, Thomashow LS, Allen AE, Paulsen IT. Comparative genomics of plant-associated Pseudomonas spp.: insights into diversity and inheritance of traits involved in multitrophic interactions. PLoS Genet. 2012;8(7):e1002784. doi: 10.1371/journal.pgen.1002784. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

