Sequencing of mRNA identifies re-expression of fetal splice variants in cardiac hypertrophy
- PMID: 23688780
- PMCID: PMC3735658
- DOI: 10.1016/j.yjmcc.2013.05.004
Sequencing of mRNA identifies re-expression of fetal splice variants in cardiac hypertrophy
Abstract
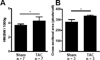
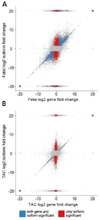
Cardiac hypertrophy has been well-characterized at the level of transcription. During cardiac hypertrophy, genes normally expressed primarily during fetal heart development are re-expressed, and this fetal gene program is believed to be a critical component of the hypertrophic process. Recently, alternative splicing of mRNA transcripts has been shown to be temporally regulated during heart development, leading us to consider whether fetal patterns of splicing also reappear during hypertrophy. We hypothesized that patterns of alternative splicing occurring during heart development are recapitulated during cardiac hypertrophy. Here we present a study of isoform expression during pressure-overload cardiac hypertrophy induced by 10 days of transverse aortic constriction (TAC) in rats and in developing fetal rat hearts compared to sham-operated adult rat hearts, using high-throughput sequencing of poly(A) tail mRNA. We find a striking degree of overlap between the isoforms expressed differentially in fetal and pressure-overloaded hearts compared to control: forty-four percent of the isoforms with significantly altered expression in TAC hearts are also expressed at significantly different levels in fetal hearts compared to control (P<0.001). The isoforms that are shared between hypertrophy and fetal heart development are significantly enriched for genes involved in cytoskeletal organization, RNA processing, developmental processes, and metabolic enzymes. Our data strongly support the concept that mRNA splicing patterns normally associated with heart development recur as part of the hypertrophic response to pressure overload. These findings suggest that cardiac hypertrophy shares post-transcriptional as well as transcriptional regulatory mechanisms with fetal heart development.
Keywords: Alternative splicing; Cardiac hypertrophy; Heart development; RNAseq.
Copyright © 2013 Elsevier Ltd. All rights reserved.
Figures




References
-
- Frey N, Olson EN. Cardiac hypertrophy: the good, the bad, and the ugly. Annu Rev Physiol. 2003;65:45–79. - PubMed
-
- Rajabi M, Kassiotis C, Razeghi P, Taegtmeyer H. Return to the fetal gene program protects the stressed heart: a strong hypothesis. Heart Fail Rev. 2007;12:331–343. - PubMed
-
- Gorza L, Pauletto P, Pessina AC, Sartore S, Schiaffino S. Isomyosin distribution in normal and pressure-overloaded rat ventricular myocardium. An immunohistochemical study. Circ Res. 1981;49:1003–1009. - PubMed
-
- Mahdavi V, Lompre AM, Chambers AP, Nadal-Ginard B. Cardiac myosin heavy chain isozymic transitions during development and under pathological conditions are regulated at the level of mRNA availability. Eur Heart J. 1984;5(Suppl F):181–191. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

