Single cell analysis of RNA-mediated histone H3.3 recruitment to a cytomegalovirus promoter-regulated transcription site
- PMID: 23689370
- PMCID: PMC3707690
- DOI: 10.1074/jbc.M113.473181
Single cell analysis of RNA-mediated histone H3.3 recruitment to a cytomegalovirus promoter-regulated transcription site
Abstract
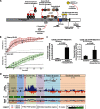
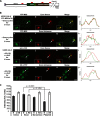
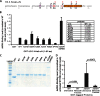
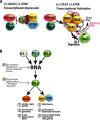
Unlike the core histones, which are incorporated into nucleosomes concomitant with DNA replication, histone H3.3 is synthesized throughout the cell cycle and utilized for replication-independent (RI) chromatin assembly. The RI incorporation of H3.3 into nucleosomes is highly conserved and occurs at both euchromatin and heterochromatin. However, neither the mechanism of H3.3 recruitment nor its essential function is well understood. Several different chaperones regulate H3.3 assembly at distinct sites. The H3.3 chaperone, Daxx, and the chromatin-remodeling factor, ATRX, are required for H3.3 incorporation and heterochromatic silencing at telomeres, pericentromeres, and the cytomegalovirus (CMV) promoter. By evaluating H3.3 dynamics at a CMV promoter-regulated transcription site in a genetic background in which RI chromatin assembly is blocked, we have been able to decipher the regulatory events upstream of RI nucleosomal deposition. We find that at the activated transcription site, H3.3 accumulates with sense and antisense RNA, suggesting that it is recruited through an RNA-mediated mechanism. Sense and antisense transcription also increases after H3.3 knockdown, suggesting that the RNA signal is amplified when chromatin assembly is blocked and attenuated by nucleosomal deposition. Additionally, we find that H3.3 is still recruited after Daxx knockdown, supporting a chaperone-independent recruitment mechanism. Sequences in the H3.3 N-terminal tail and αN helix mediate both its recruitment to RNA at the activated transcription site and its interaction with double-stranded RNA in vitro. Interestingly, the H3.3 gain-of-function pediatric glioblastoma mutations, G34R and K27M, differentially affect H3.3 affinity in these assays, suggesting that disruption of an RNA-mediated regulatory event could drive malignant transformation.
Keywords: Antisense RNA; Chromatin Regulation; Chromatin Remodeling; Gene Expression; Gene Regulation; Gene Silencing; Microscopy; Nuclear Organization; RNA; Transcription.
Figures





Similar articles
-
RNase P protein subunit Rpp29 represses histone H3.3 nucleosome deposition.Mol Biol Cell. 2016 Apr 1;27(7):1154-69. doi: 10.1091/mbc.E15-02-0099. Epub 2016 Feb 3. Mol Biol Cell. 2016. PMID: 26842893 Free PMC article.
-
H3.Y discriminates between HIRA and DAXX chaperone complexes and reveals unexpected insights into human DAXX-H3.3-H4 binding and deposition requirements.Nucleic Acids Res. 2017 Jun 2;45(10):5691-5706. doi: 10.1093/nar/gkx131. Nucleic Acids Res. 2017. PMID: 28334823 Free PMC article.
-
Single-cell analysis of Daxx and ATRX-dependent transcriptional repression.J Cell Sci. 2012 Nov 15;125(Pt 22):5489-501. doi: 10.1242/jcs.110148. Epub 2012 Sep 12. J Cell Sci. 2012. PMID: 22976303 Free PMC article.
-
All roads lead to chromatin: multiple pathways for histone deposition.Biochim Biophys Acta. 2013 Mar-Apr;1819(3-4):238-46. Biochim Biophys Acta. 2013. PMID: 24459726 Review.
-
Histone H3 variants specify modes of chromatin assembly.Proc Natl Acad Sci U S A. 2002 Dec 10;99 Suppl 4(Suppl 4):16477-84. doi: 10.1073/pnas.172403699. Epub 2002 Aug 12. Proc Natl Acad Sci U S A. 2002. PMID: 12177448 Free PMC article. Review.
Cited by
-
A scoping review of diffuse hemispheric glioma, H3 G34-mutant: Epigenetic and molecular profiles, clinicopathology, and treatment avenues.Neurooncol Adv. 2024 Dec 7;6(1):vdae208. doi: 10.1093/noajnl/vdae208. eCollection 2024 Jan-Dec. Neurooncol Adv. 2024. PMID: 39759262 Free PMC article.
-
Reshaping chromatin after DNA damage: the choreography of histone proteins.J Mol Biol. 2015 Feb 13;427(3):626-36. doi: 10.1016/j.jmb.2014.05.025. Epub 2014 Jun 2. J Mol Biol. 2015. PMID: 24887097 Free PMC article. Review.
-
ATRX promotes maintenance of herpes simplex virus heterochromatin during chromatin stress.Elife. 2018 Nov 22;7:e40228. doi: 10.7554/eLife.40228. Elife. 2018. PMID: 30465651 Free PMC article.
-
PML nuclear bodies and chromatin dynamics: catch me if you can!Nucleic Acids Res. 2020 Dec 2;48(21):11890-11912. doi: 10.1093/nar/gkaa828. Nucleic Acids Res. 2020. PMID: 33068409 Free PMC article. Review.
-
An evolving view of epigenetic complexity in the brain.Philos Trans R Soc Lond B Biol Sci. 2014 Sep 26;369(1652):20130506. doi: 10.1098/rstb.2013.0506. Philos Trans R Soc Lond B Biol Sci. 2014. PMID: 25135967 Free PMC article. Review.
References
-
- Talbert P. B., Henikoff S. (2010) Histone variants. Ancient wrap artists of the epigenome. Nat. Rev. Mol. Cell Biol. 11, 264–275 - PubMed
-
- Waterborg J. H. (2012) Evolution of histone H3. Emergence of variants and conservation of post-translational modification sites. Biochem. Cell Biol. 90, 79–95 - PubMed
-
- Ahmad K., Henikoff S. (2002) The histone variant H3.3 marks active chromatin by replication-independent nucleosome assembly. Mol. Cell 9, 1191–1200 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

