Dismantling promoter-driven RNA polymerase II transcription complexes in vitro by the termination factor Rat1
- PMID: 23689372
- PMCID: PMC3707679
- DOI: 10.1074/jbc.M112.434985
Dismantling promoter-driven RNA polymerase II transcription complexes in vitro by the termination factor Rat1
Abstract
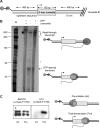
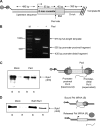
Proper RNA polymerase II (Pol II) transcription termination is essential to generate stable transcripts, to prevent interference at downstream loci, and to recycle Pol II back to the promoter (1-3). As such, termination is an intricately controlled process that is tightly regulated by a variety of different cis- and trans-acting factors (4, 5). Although many eukaryotic termination factors have been identified to date, the details of the precise molecular mechanisms governing termination remain to be elucidated. We devised an in vitro transcription system to study specific Pol II termination. We show for the first time that the exonucleolytic Rat1·Rai1 complex can elicit the release of stalled Pol II in vitro and can do so in the absence of other factors. We also find that Rtt103, which interacts with the Pol II C-terminal domain (CTD) and with Rat1, can rescue termination activity of an exonucleolytically deficient Rat1 mutant. In light of our findings, we posit a model whereby functional nucleolytic activity is not the feature of Rat1 that ultimately promotes termination. Degradation of the nascent transcript allows Rat1 to pursue Pol II in a guided fashion and arrive at the site of RNA exit from Pol II. Upon this arrival, however, it is perhaps the specific and direct contact between Rat1 and Pol II that transmits the signal to terminate transcription.
Keywords: Enzyme Mechanisms; Molecular Biology; RNA Polymerase II; Rat1; Transcription Termination; Yeast Transcription.
Figures


Similar articles
-
Unraveling the mechanistic features of RNA polymerase II termination by the 5'-3' exoribonuclease Rat1.Nucleic Acids Res. 2015 Mar 11;43(5):2625-37. doi: 10.1093/nar/gkv133. Epub 2015 Feb 26. Nucleic Acids Res. 2015. PMID: 25722373 Free PMC article.
-
Molecular basis for the interaction between Saccharomyces cerevisiae Rtt103 and the Rat1-Rai1 complex.Nat Commun. 2025 Apr 5;16(1):3266. doi: 10.1038/s41467-025-58671-z. Nat Commun. 2025. PMID: 40188244 Free PMC article.
-
Torpedo nuclease Rat1 is insufficient to terminate RNA polymerase II in vitro.J Biol Chem. 2009 Aug 7;284(32):21270-9. doi: 10.1074/jbc.M109.013847. Epub 2009 Jun 17. J Biol Chem. 2009. PMID: 19535338 Free PMC article.
-
Transcription termination and the control of the transcriptome: why, where and how to stop.Nat Rev Mol Cell Biol. 2015 Mar;16(3):190-202. doi: 10.1038/nrm3943. Epub 2015 Feb 4. Nat Rev Mol Cell Biol. 2015. PMID: 25650800 Review.
-
Genome-wide RNA polymerase II: not genes only!Trends Biochem Sci. 2008 Jun;33(6):265-73. doi: 10.1016/j.tibs.2008.04.006. Epub 2008 May 6. Trends Biochem Sci. 2008. PMID: 18467100 Review.
Cited by
-
RNA Polymerase II CTD phosphatase Rtr1 fine-tunes transcription termination.PLoS Genet. 2020 Mar 18;16(3):e1008317. doi: 10.1371/journal.pgen.1008317. eCollection 2020 Mar. PLoS Genet. 2020. PMID: 32187185 Free PMC article.
-
Genome-wide mapping of yeast RNA polymerase II termination.PLoS Genet. 2014 Oct 9;10(10):e1004632. doi: 10.1371/journal.pgen.1004632. eCollection 2014 Oct. PLoS Genet. 2014. PMID: 25299594 Free PMC article.
-
Effects of Transcription Elongation Rate and Xrn2 Exonuclease Activity on RNA Polymerase II Termination Suggest Widespread Kinetic Competition.Mol Cell. 2015 Oct 15;60(2):256-67. doi: 10.1016/j.molcel.2015.09.026. Mol Cell. 2015. PMID: 26474067 Free PMC article.
-
Writing a wrong: Coupled RNA polymerase II transcription and RNA quality control.Wiley Interdiscip Rev RNA. 2019 Jul;10(4):e1529. doi: 10.1002/wrna.1529. Epub 2019 Mar 7. Wiley Interdiscip Rev RNA. 2019. PMID: 30848101 Free PMC article. Review.
-
Application of the SSB biosensor to study in vitro transcription.Biochem Biophys Res Commun. 2018 Feb 12;496(3):820-825. doi: 10.1016/j.bbrc.2018.01.147. Epub 2018 Jan 31. Biochem Biophys Res Commun. 2018. PMID: 29378185 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

