Analysis of polymorphic residues reveals distinct enzymatic and cytotoxic activities of the Streptococcus pyogenes NAD+ glycohydrolase
- PMID: 23689507
- PMCID: PMC3707703
- DOI: 10.1074/jbc.M113.481556
Analysis of polymorphic residues reveals distinct enzymatic and cytotoxic activities of the Streptococcus pyogenes NAD+ glycohydrolase
Abstract
The Streptococcus pyogenes NAD(+) glycohydrolase (SPN) is secreted from the bacterial cell and translocated into the host cell cytosol where it contributes to cell death. Recent studies suggest that SPN is evolving and has diverged into NAD(+) glycohydrolase-inactive variants that correlate with tissue tropism. However, the role of SPN in both cytotoxicity and niche selection are unknown. To gain insight into the forces driving the adaptation of SPN, a detailed comparison of representative glycohydrolase activity-proficient and -deficient variants was conducted. Of a total 454 amino acids, the activity-deficient variants differed at only nine highly conserved positions. Exchanging residues between variants revealed that no one single residue could account for the inability of the deficient variants to cleave the glycosidic bond of β-NAD(+) into nicotinamide and ADP-ribose; rather, reciprocal changes at 3 specific residues were required to both abolish activity of the proficient version and restore full activity to the deficient variant. Changing any combination of 1 or 2 residues resulted in intermediate activity. However, a change to any 1 residue resulted in a significant decrease in enzyme efficiency. A similar pattern involving multiple residues was observed for comparison with a second highly conserved activity-deficient variant class. Remarkably, despite differences in glycohydrolase activity, all versions of SPN were equally cytotoxic to cultured epithelial cells. These data indicate that the glycohydrolase activity of SPN may not be the only contribution the toxin has to the pathogenesis of S. pyogenes and that both versions of SPN play an important role during infection.
Keywords: Allelic Variation; Bacteria; Bacterial Pathogenesis; Bacterial Toxins; Cell Death; Cytotoxicity; Enzymes; Group A Streptococcus; NAD+ Glycohydrolase.
Figures

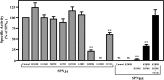
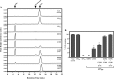


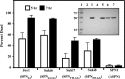

References
-
- Madden J. C., Ruiz N., Caparon M. (2001) Cytolysin-mediated translocation (CMT): a functional equivalent of type III secretion in Gram-positive bacteria. Cell 104, 143–152 - PubMed
-
- Ghosh J., Caparon M. G. (2006) Specificity of Streptococcus pyogenes NAD+ glycohydrolase in cytolysin-mediated translocation. Mol. Microbiol. 62, 1203–1214 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

